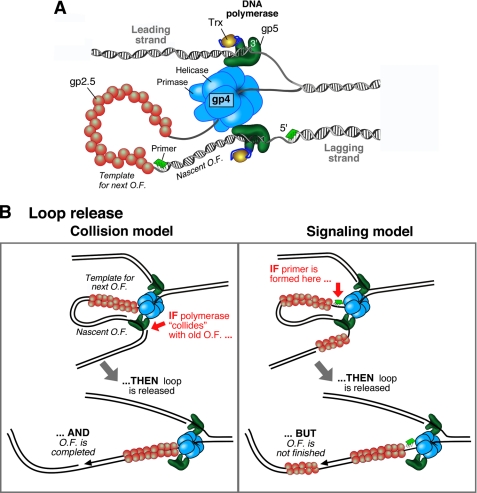
FIGURE 1.
A, organization of the bacteriophage T7 replication fork. gp4 encircles the lagging strand and mediates both the unwinding of dsDNA via its helicase domain and the synthesis of RNA primers via the primase domain. The T7 DNA polymerases are stably bound to gp4 and incorporate nucleotides on the leading and lagging strands. The DNA polymerase is a 1:1 complex of T7 gp5 and E. coli Trx. The ssDNA extruded behind the helicase is coated by the ssDNA-binding protein gp2.5. A replication loop is formed in the lagging strand to align it with the leading strand. The lagging-strand DNA polymerase initiates Okazaki fragment (O.F.) synthesis using RNA primers (green segments). B, schematic depiction of the two models that describe replication loop release. In the collision model (left panel), the replication loop is released when the lagging-strand polymerase collides with the 5′ terminus of the previous Okazaki fragment. In the signaling model (right panel), the synthesis of a new primer triggers the release of the replication loop prior to the completion of the nascent Okazaki fragment.