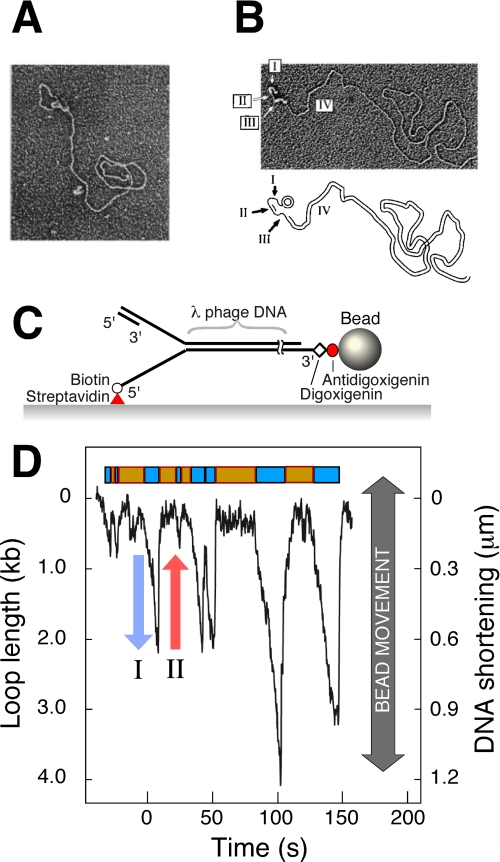
FIGURE 2.
A, observation of replication loops by EM. Coordinated DNA synthesis is carried out on a 70-bp minicircle substrate. The replication proteins appear as a globular object. The dsDNA loop extending from the replisome represents the nascent Okazaki fragment. Both the small minicircle and the condensed gp2.5-ssDNA complex are obscured by the protein mass containing the replisome. The figure is from Ref. 22. B, observation of Okazaki fragments by EM. The replication reaction was carried out as described for A, deproteinated, and treated with SSB. SSB extends ssDNA and enables its visualization as a DNA segment with increased diameter. Segment I indicates the template for the next Okazaki fragment produced by the helicase, segment II is the nascent Okazaki fragment, segment III is the template of the nascent Okazaki fragment, and segment IV is the long dsDNA of previously synthesized Okazaki fragments. The figure is from Ref. 22. C, flow-stretching individual DNA molecules. Duplex λ-DNA (48.5 kb) is modified to contain a replication fork. The lagging strand of the forked end is coupled to the surface. The other end of the DNA is attached to a bead. A constant laminar flow applies a well controlled drag force to the bead and stretches the DNA molecule. The length of the individual DNA molecules is measured by imaging the beads and tracking their positions. The figure is from Ref. 38. D, dynamic single-molecule observation of replication loops produced by individual replisomes. The trajectory shows a time course of the length of a single DNA molecule during replication. The DNA shortening corresponds to loop growth during leading- and lagging-strand synthesis (blue arrow) and is followed by a rapid length increase when a loop is released (red arrow). The lag phases between looping events and the loop growth phases are shown as orange and cyan boxes, respectively. The figure is from Ref. 38.