FIGURE 6.
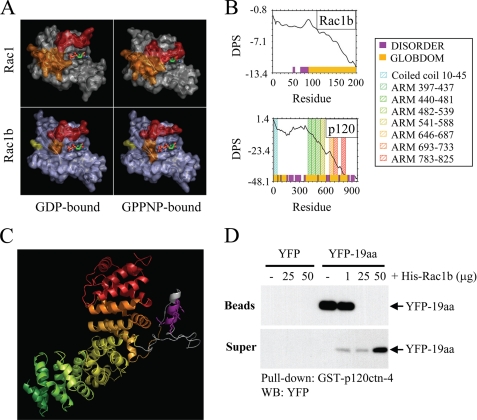
p120ctn and Rac1b bind through the interaction of protein regions predicted to be disordered. A, most protein interactions of Rac1 (top panels) involve binding through switch 1 (red) and/or switch 2 (orange); the conformations of these switches are determined by binding to GDP (top left) or GTP (mimicked by uncleavable analog GPPNP; top right). In Rac1b (lower panels), switch 1 (red) fails to adopt a closed conformation upon GPPNP binding. B, disorder propensity sum (DPS) plots predict intrinsically unstructured regions for Rac1b (left) and p120ctn (right). Regions of disorder, characterized by a positive slope on the plot, are represented with magenta blocks along the x axis, whereas predicted globular domains, characterized by negative slope, are represented with gold blocks. Residues 607–644 of p120ctn (containing the polylysine motif) and residues 70–90 of Rac1b (encompassing much of the 19-aa insertion) are predicted to be unstructured. C, homology model of the ARM domain of p120ctn shows that the polylysine motif (magenta) is located in an insertion between ARM repeats 5 and 6. ARM repeats are colored to correspond to the Pfam domains indicated in the plot above. The polylysine motif (magenta) and flanking sequences shown in gray correspond to disordered regions in the crystal structure of plakophilin 1, upon which the homology model is based. D, cells were transfected with YFP or YFP-19aa, and lysates were precipitated with glutathione beads or were preincubated with 1, 25, or 50 μg of His-Rac1b prior to precipitation. Bead precipitates and supernatants were solubilized, Western blotted (WB), and probed with anti-YFP antibodies.