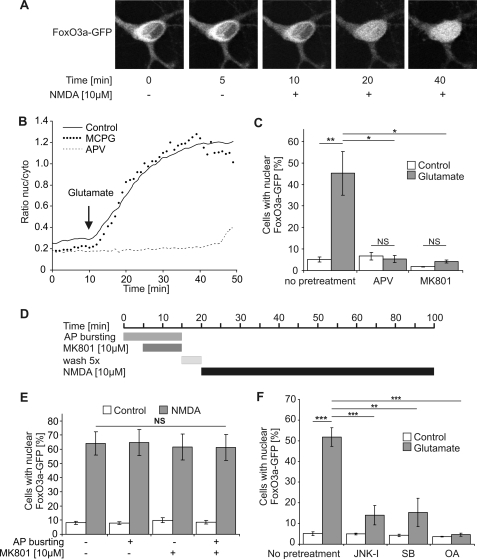
FIGURE 2.
Role of extrasynaptic NMDA receptors, JNK, p38 MAP kinase, and protein phosphatase 2A in glutamate/NMDA-induced nuclear translocation of FoxO3a-GFP. A, live imaging of FoxO3a-GFP in hippocampal neurons before and after treatment with 10 μm NMDA is shown. The confocal images are z-stacks projected into one plane and were acquired at the indicated times before and after NMDA application. A representative example is shown. B, shown is live imaging of FoxO3a-GFP in hippocampal neurons before and after treatment with 10 μm glutamate in the absence of glutamate receptor blockers (solid line) or in the presence of 100 μm APV (dashed line) or 500 μm MCPG (dotted line). The arrow indicates the time point of glutamate application. The traces shown represent the average ratios of the nuclear and cytoplasmic FoxO3a-GFP signals obtained from four neurons (Control), five neurons (APV), and three neurons (MCPG). nuc/cyto, nucleo-cytoplasmic. C, F, shown is the quantitative analysis of the percentage of hippocampal neurons with nuclear localized FoxO3a-GFP. Unstimulated hippocampal neurons (Control) and hippocampal neurons stimulated for 1 h with 10 μm glutamate were pretreated with 100 μm APV, 10 μm MK-801, 20 μm JNK-I peptide, 10 μm SB203580 (SB), and 0.2 nm okadaic acid (OA) or received no pretreatment (n = 3). Number of cells analyzed per condition (i.e. with and without glutamate stimulation): 2331, no pretreatment; 2120, APV; 2643, MK-801; 2014, JNK-1; 1963, SB203580; 2713, okadaic acid. Statistically significant differences (ANOVA followed by Tukey's post hoc test) are indicated with asterisks; ***, p < 0.001; **, p < 0.01; *, p < 0.05. Bars represent the means ± S.E. (n = 4). NS, not significant. D, shown is a schematic illustration of the protocol for selective stimulation of extrasynaptic NMDA receptors. E, shown is a quantitative analysis of the percentage of hippocampal neurons with nuclear localized FoxO3a-GFP. Neurons did or did not receive pretreatment with bicuculline/MK-801 to isolate extrasynaptic NMDA receptors and were subsequently exposed for 1 h to 10 μm NMDA. For each condition in each experiment about 300 cells were analyzed. Statistical analysis was determined by ANOVA followed by Tukey's post hoc test; NS, not significant. Bars represent the means ± S.E. (n = 5).