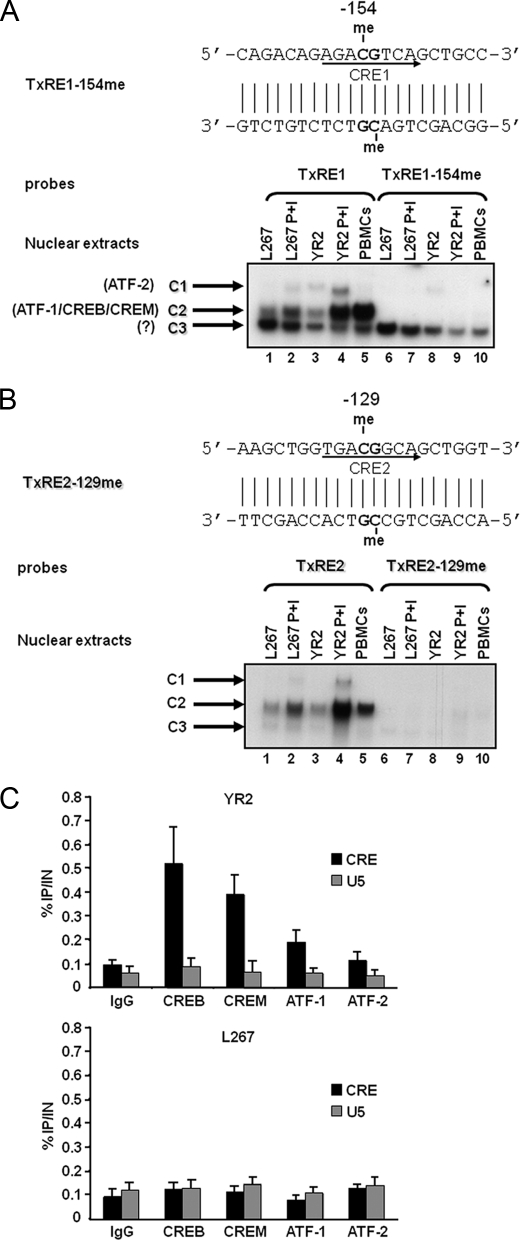
FIGURE 6.
Inhibition of CREB/CREM/ATF-1 transcription factor binding by methylation at the −154 and −129 CpGs. A, EMSA analysis of nuclear factors interacting with the methylated or unmethylated TxRE1 probes. The nucleotide sequence of the methylated TxRE1–154me double-stranded oligonucleotide is shown with the CRE1 site indicated by an arrow on the coding strand and with the methylated cytosine residues indicated by the abbreviation me. The unmethylated TxRE1 double-stranded oligonucleotide (lanes 1–5) or its methylated version, TxRE1–154me (lanes 6–10), was used as probe and incubated with nuclear extracts from the BLV latently infected B-cell line L267 (30 μg) treated (lanes 2 and 7) or not (lanes 1 and 6) with PMA + ionomycin (P+I) for 30 min, from the defective cell line YR2 (15 μg) treated (lanes 4 and 9) or not (lanes 3 and 8) with PMA + ionomycin, or from BLV-infected ovine PBMCs (10 μg) (lanes 5 and 10). Sequence-specific retarded bands of interest are shown. A minor and two major protein-DNA complexes were observed and named C1, C2, and C3, respectively (indicated by arrows). The presence of ATF-2 in the minor protein-DNA complex C1 and the presence of ATF-1, CREB, and CREM in the major protein-DNA complex C2 were reported previously by our laboratory (17). B, EMSA analysis of nuclear factors interacting with the methylated or unmethylated TxRE2 probes. The nucleotide sequence of the methylated TxRE2–129me double-stranded oligonucleotide is shown with the CRE2 site indicated by an arrow on the coding strand and with the methylated cytosine residues indicated by the abbreviation me. The unmethylated TxRE2 double-stranded oligonucleotide (lanes 1–5) or its methylated version, TxRE2–129me (lanes 6–10), was used as probe and incubated with nuclear extracts from the BLV latently infected B-cell line L267 (30 μg) treated (lanes 2 and 7) or not (lanes 1 and 6) with PMA + ionomycin for 30 min; from the defective cell line YR2 (15 μg) treated (lanes 4 and 9) or not (lanes 3 and 8) with PMA + ionomycin; or from BLV-infected ovine PBMCs (10 μg) (lanes 5 and 10). Sequence-specific retarded bands of interest are shown. A minor and two major protein-DNA complexes were observed (C1, C2, and C3, respectively; indicated by arrows). The presence of ATF-2 in the minor protein-DNA complex C1 and the presence of ATF-1, CREB, and CREM in the major protein-DNA complex C2 were reported previously by our laboratory (17). C, CREB/CREM/ATF-1 are recruited in vivo to the BLV 5′-LTR CRE region. YR2 and L267 cells were cross-linked for 10 min at room temperature with 1% formaldehyde. To detect chromosomal flanking regions, pellets were sonicated to obtain DNA fragments of an average size of 400 bp. Chromatin immunoprecipitations were performed with an antibody directed against CREB, CREM, ATF-1, or ATF-2. To test aspecific binding to the beads, a purified IgG was used as a control for immunoprecipitation. Quantitative PCR reactions were performed with oligonucleotide primers hybridizing in a region overlapping the three CRE sites of the BLV LTR or in the U5 region of the BLV LTR, where no CRE binding site has been reported previously. -Fold enrichments were calculated as the percentage of input values following the formula: Immunoprecipitated DNA (IP)/total DNA (IN). Values represent the means of triplicate samples ± S.E. An experiment representative of three independent ChIP assays is shown.