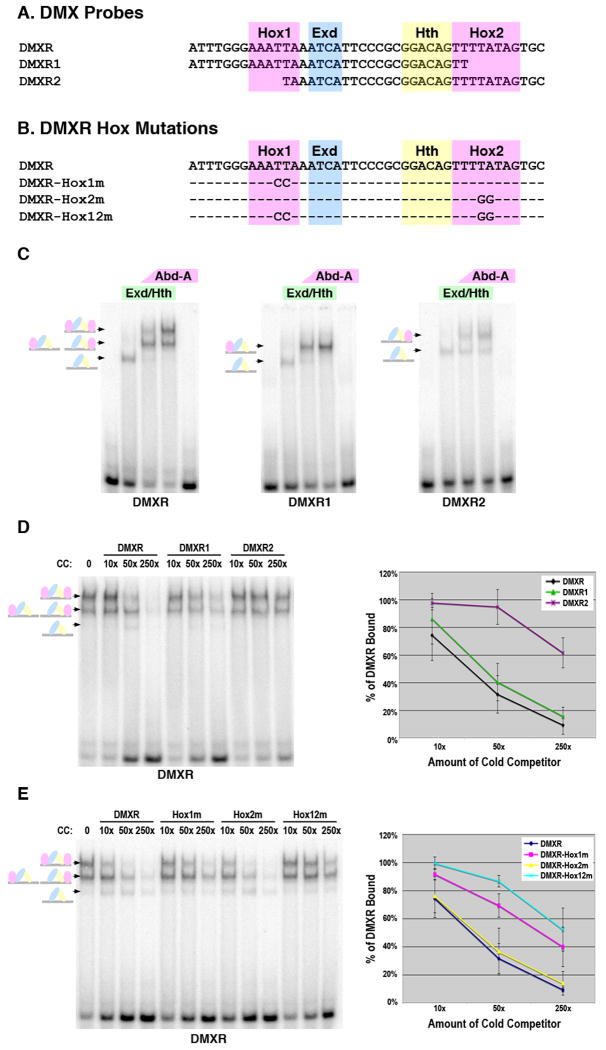
Figure 2. Dependence of Abd-A tetramer formation on the presence of two Hox sites within DMXR.
A. The DMXR probes used in EMSAs with the Hox1, Exd, Hth, and Hox2 sites highlighted. B. The DMXR Hox point mutations used in gel shift analysis. C. Comparative EMSAs of Hox complex formation on the DMXR, DMXR1, and DMXR2 probes. Conditions are as follows: First lane is probe alone, second lane is 75 × 10-9 M of Exd/Hth heterodimers, third and fourth lane contain the same amount of Exd/Hth with either 17 × 10-9 M (low amount) or 75 × 10-9 M (high amount) of Abd-A, respectively and the fifth lane contains 75 × 10-9 M of Abd-A alone. Schematics at left denote color-coded complexes formed (Exd, blue; Hth, yellow; Abd-A, pink). D and E. Competition DNA binding assays for Abd-A/Exd/Hth complexes on labeled DMXR. Each lane contains 75 × 10-9 M of Exd/Hth and Abd-A. The first lane contains no competitor DNA whereas subsequent lanes contain either 10×, 50×, or 250× of the indicated cold competitor. Schematics at left denote complexes (Exd, blue; Hth, yellow; Abd-A, pink). Graph depicts the average percent of DMXR probe bound from three different experiments in the presence of different amounts of each cold competitor probe. For this analysis, the amount of probe bound was determined using phosphor-imaging densitometry, and 100% binding was assigned to the amount of probe bound in the absence of competitor. Standard error is noted.