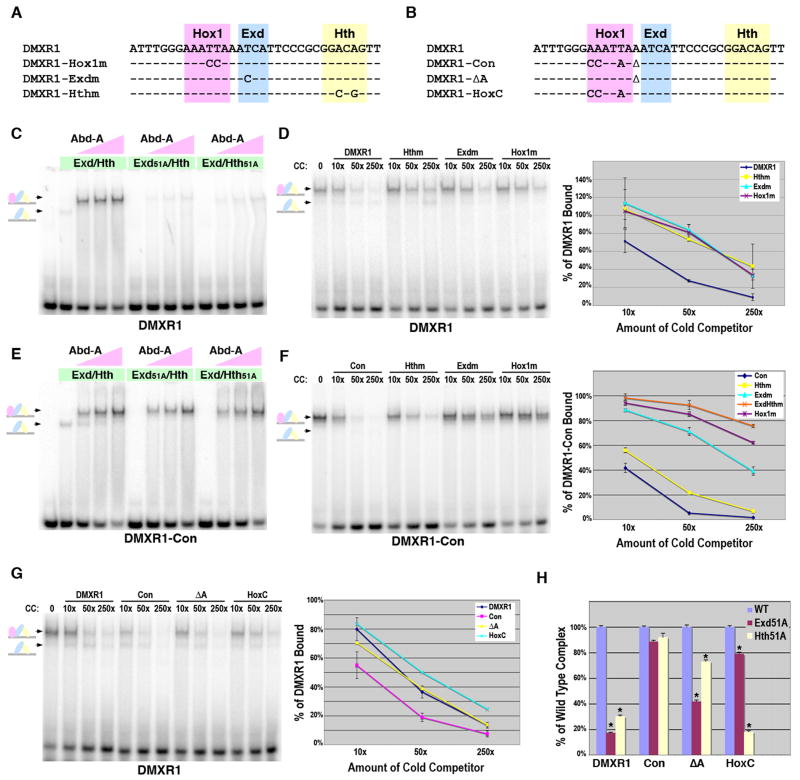
Figure 4. Role of Hth binding for Hox complex formation on Exd/Hox sites.
A. The DMXR1 Hox1, Exd, and Hth point mutations used in gel shift analysis. B. Sequence comparisons of the DMXR1, DMXR1-Con, DMXR1-ΔA, and DMXR1-HoxC probes used in gel shift assays. C. Comparative EMSAs using wild type and mutant Exd/Hth heterodimers with Abd-A on DMXR1. Equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) were used with three concentrations of Abd-A (7.5 × 10-9 M, 22.5 × 10-9 M, and 70 × 10-9 M). D. Dependence of Abd-A/Exd/Hth binding to DMXR1 on the Hox1, Exd, and Hth sites. DNA competition assays were performed using labeled DMXR1 and DMXR1, Hox1m, Exdm, and Hthm probes as cold competitors. The amount of probe bound in absence of competitor was assigned 100% binding and the graph depicts the average amount of probe bound in presence of each competitor from three different experiments. E. Comparative EMSAs using wild type and mutant Exd/Hth heterodimers with Abd-A on DMXR1-Con. Equimolar amounts of Exd/Hth, Exd51A/Hth, and Exd/Hth51A proteins (30 × 10-9 M) were used with three amounts of Abd-A (7.5 × 10-9 M, 22.5 × 10-9 M, and 70 × 10-9 M). F. Dependence of Abd-A/Exd/Hth binding to DMXR1-Con on the Hox1, Exd, and Hth sites. DNA binding competition assays were performed using labeled DMXR1-Con and different amounts of DMXR1-Con wild type, Hox1m, Exdm, Hthm, and ExdmHthm probes as cold competitors. The amount of probe bound in absence of competitor was assigned 100% binding and the graph depicts the average amount of probe bound in presence of competitor from three different experiments. G. DNA binding competition assays of Abd-A, Exd, and Hth complex formation on the DMXR1, DMXR1-Con, DMXR1-ΔA, and DMXR1-HoxC probes. Labeled DMXR1 probe was bound with a constant amount of Exd/Hth and Abd-A. Different amounts of competitor were added as indicated. Schematics at left denote color-coded complexes (Exd, blue; Hth, yellow; Abd-A, pink). Data from three independent experiments is graphed at right. Note only the DMXR1-Con is significantly different from the wild type DMXR1 (p-value < 0.01) H. Assessing the dependence of Hox complex formation on Exd and Hth binding to DMXR1, DMXR1-Con, DMXR1-ΔA, and DMXR1-HoxC. Comparative DNA binding assays were performed in triplicate using equimolar amounts of Exd/Hth, Exd51A/Hth, or Exd/Hth51A proteins and Abd-A. The amount of probe bound by Abd-A and wild type Exd/Hth was assigned to 100% for each probe tested (blue bar). The amount of probe bound by Exd51A/Hth (red bar) and Exd/Hth51A (yellow bar) compared to wild type was then determined. * denotes a significant difference from wild type binding (p-value < 0.001).