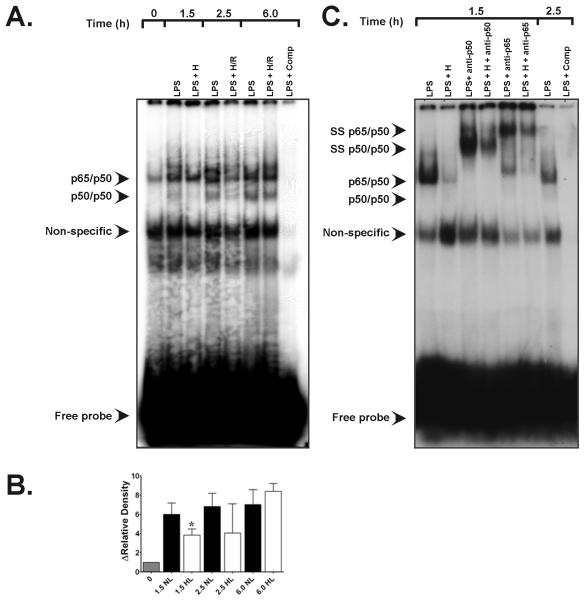
Figure 4.
A. Representative EMSA specific for NF-κB transactivation in rat alveolar macrophages showing the effects of hypoxic co-stimulation on E. coli serotype O55:B5 LPS-induced NF-κB DNA-binding activity. Bands of the NF-κB p65/p50 heterodimeric complex and of p50/p50 homodimers are depicted. Compared with normoxic LPS controls (90NL), 90 min of hypoxia (H) starting after LPS treatment at t = 0 (1.5 h) suppressed heterodimeric NF-κB activity as well as p50/p50 homodimeric signal. Lane 1, t = 0 baseline control without LPS or hypoxic co-stimulation; lane 2, signal from normoxic LPS control obtained 1.5 h after E. coli LPS stimulation (see Materials and Methods); lane 3, time-matched signal for LPS + H at peak hypoxia; lane 4, normoxic LPS control signal at 2.5 h; lane 5, time-matched signal for LPS + H/R cells at 2.5 h; lane 6, normoxic LPS control 6.0 h after initial LPS stimulation; lane 7, time-matched LPS + H/R at 6.0 h ; lane 8, (LPS + comp), competition of an LPS sample with excess unlabeled NF-κB oligonucleotide. B. Group-specific densitometric data (means ± SE) from the EMSA shown in panel A above, representing at least three separate experiments for each time point. * P < 0.05 vs. time-matched 1.5 h LPS normoxic LPS control. C. Representative EMSA depicting the results of supershift analyses for the NF-κB heterodimeric complex. Lane 1, signal from normoxic LPS control lysates obtained 1.5 h after E. coli LPS stimulation; lane 2, time-matched signal from LPS + hypoxia cell lysates at t = 1.5 h; lane 3, effects of anti-p65 antibody (Ab) treatment on normoxic LPS signal at t = 1.5 h; lane 4, effects of anti-p65 Ab treatment on LPS + 1.5 h hypoxia lysates; lane 5, effects of anti-p50 Ab treatment on normoxic LPS lysates at t = 1.5 h; lane 6, signal from t = 2.5 h normoxic LPS controls; lane 7, competition with excess unlabeled NF-κB oligonucleotide.