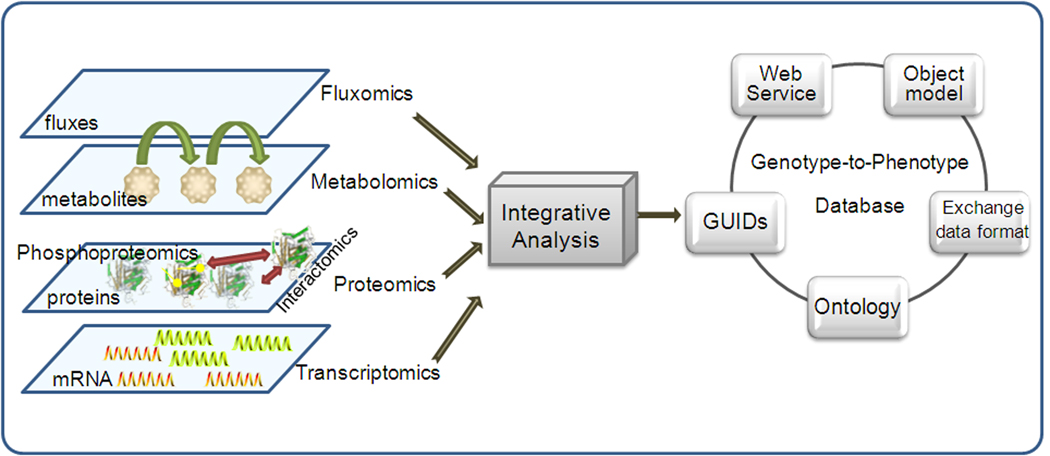
Figure 3. The conceptual work flow of integrated analysis of –omics data to formalize genotype-to-phenotype database.
Advances in -omics measurement lead to the accumulation of high throughput -omics data. With systematic and integrative analysis tools, genotype-to-phenotype database are currently being developed under the core modules outlined in this figure. Standardized object models and data formats facilitate the exchange of information between different data systems and are essential for unambiguous transmission of data between computers. A well defined ontology enables the representation of domain-specific knowledge, as well as, the relationship between those domains and leads to powerful database searching. GUIDs solve data integration problems that result from ambiguity in name, or identity, of biological concepts and objects. Standard protocols for web service afford machine-to-machine interaction over the internet and simplify the task of exploring distributed data, forming the basis of the service-oriented architecture (SOA) [44].