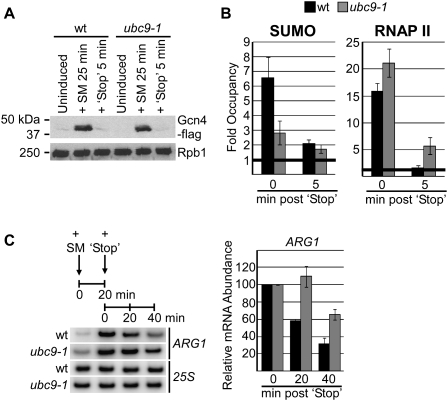
Figure 5.
Sumoylation is required for deactivation of the induced ARG1 gene. (A) ubc9-1 and isogenic wild-type cells transformed with pGcn4-Flag plasmid were grown at 28°C, and cell lysates were prepared from aliquots removed prior to induction (Uninduced), 25 min after adding SM, then 5 min after adding stop mix consisting of fivefold concentrated Val and Ile. Western blot is shown of Gcn4-Flag expression using a Flag antibody and 8WG16 antibody for loading control (Rpb1). (B) Occupancy of SUMO and RNAP II at the ARG1 promoter (position A in Fig. 4B) was determined 25 min after induction with SM, and 5 min after adding the stop mix to wild-type (black bars) and ubc9-1 (gray bars) cells. Statistical analysis of RNAP II levels associated with ARG1 after addition of the stop mix indicates that significant levels were detected in ubc9-1 cells, and no significant levels were detected in wild-type cells (P < 0.04 and P > 0.1, respectively, compared with null hypothesis of fold occupancy equal to 1). (C, left) Transcript abundance of ARG1 was determined by RT–PCR on total RNA isolated from the indicated samples at 0 and 20 min post-addition of SM, and 20 and 40 min post-addition of the stop mix. RNAP I-transcribed 25S RNA analysis is shown as a control. Quantification of three RT–PCR analyses, showing values for 0, 20 and 40 min post-addition of stop mix, is shown at right, normalized to abundance of ARG1 mRNA 20 min post-addition of SM in wild-type and ubc9-1 cells (0 min post-stop).