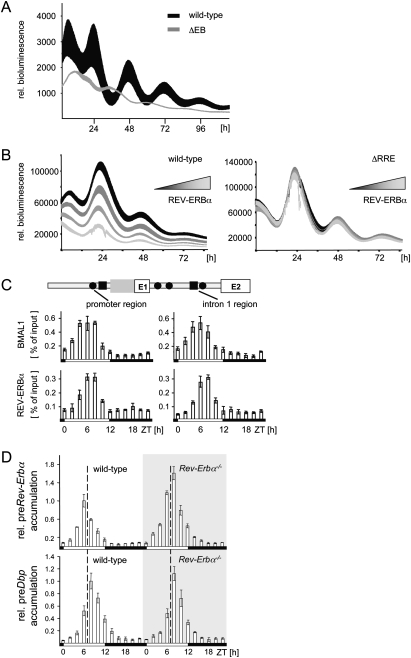
Figure 5.
Autorepression of the Rev-Erbα gene. (A) Identification of functional E-boxes within the Rev-Erbα gene. NIH 3T3 fibroblasts were transfected with a reporter gene containing the wild-type regulatory region of the Rev-Erbα gene (black), or a version with all potential E-boxes inactivated (gray). (B) Identification of functional RREs within the Rev-Erbα gene. The genomic Rev-Erbα-luciferase construct, or a version with the two potential REV-ERBα-binding sites mutated, was cotransfected with increasing amounts (22, 66, or 200 ng) of a vector containing the entire Rev-Erbα gene (average from n = 4; mean ± SD). (C) Chromatin immunoprecipitation analysis to monitor the binding of BMAL1 or REV-ERBα to the promoter region or the intron 1 region of the Rev-Erbα gene using the same chromatin as in Figure 3A. Above is a scheme of the regulatory region of the Rev-Erbα gene around exon 1 and exon 2 indicating the positions of potential E-boxes (black circles) or potential RREs (black squares). (D) Precursor RNA accumulation of Dbp or Rev-Erbα in liver tissue from wild-type or Rev-Erbα-deficient mice at 2-h resolutions. The peak of mRNA accumulation observed in the wild-type mice was arbitrarily set to 1.0 (n = 3; mean ± SD). Note that, in Rev-Erbα–deficient mice, we measure the intron 1 of the knockout allele, which is identical to the intron 1 of the Rev-Erbα gene and probably has the same half-life.