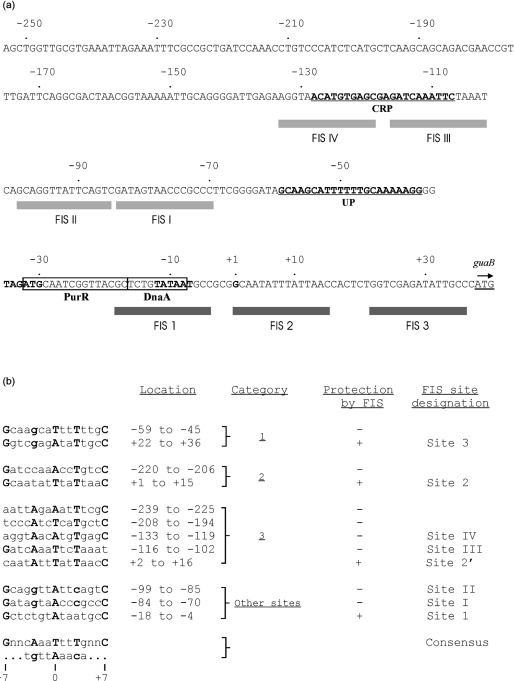
Fig. 1.
Identification of putative FIS sites at PguaB. (a) Sequence of the guaB promoter from positions −253 to +40 relative to the guaB transcription start site. Putative FIS sites I–IV, FIS sites 1–3, a putative CRP binding site centred at position −117.5 and binding sites for PurR and DnaA are indicated (Hutchings & Drabble, 2000). The core promoter elements (−35 and −10 regions), UP element, and the initiating nucleotide are also shown, in bold. Apart from FIS sites I–IV, putative FIS sites that have been shown not to bind FIS in this work are not indicated. For clarity, FIS site 2′ is not shown. (b) Candidate FIS binding sites were identified by comparing the consensus sequence for FIS (Ross et al., 1999; Shultzaberger et al., 2007) with PguaB sequences located from positions −253 to +36. The consensus sequence employed is indicated accordingly; alternative bases at each position of the consensus sequence are shown in the second line. Conserved bases at the five positions considered to be critical for FIS binding (positions −7, −3, 0, +3 and +7) are emboldened, and the most frequently occurring base is shown in upper case. ‘n' signifies any base. Candidate FIS sites are categorized according to their similarity to the consensus. Category 1 and 2 sites exhibit a 4/5 match at the critical positions, including positions +7 and −7, which are most strongly conserved among FIS sites. However, at category 2 sites, an infrequently occurring base is present at position −3, 0 or +3 (in the case of PguaB, such mismatches only occur at position −3). Category 3 sites also exhibit a 4/5 match at the critical positions but the mismatched base occurs at position +7 or −7. Other pertinent sites that do not fulfil the criteria for inclusion in categories 1–3 are shown as ‘other sites'. FIS sites that were protected by FIS in DNase I footprinting are as indicated.