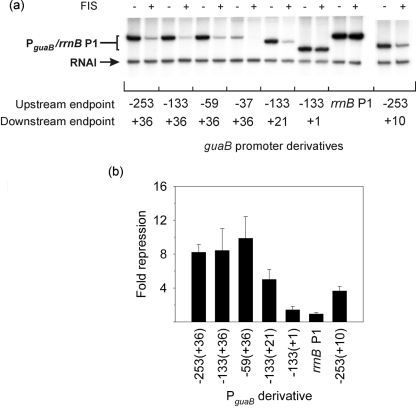
Fig. 4.
Role of FIS in regulation of transcription from PguaB in vitro. (a) Multiple-round in vitro transcription was employed to measure transcription from PguaB derivatives PguaB (-253 to +36), PguaB (-133 to +36), PguaB (-59 to +36), PguaB (-37 to +36), PguaB (-133 to +1), PguaB (-133 to +21) and PguaB (-253 to +10) in the presence (+) and absence (−) of 250 nM FIS. Promoter endpoints are as indicated. PguaB (-133 to +21) lacks FIS site 3, and PguaB (-133 to +1) lacks FIS sites 2 and 3. As a control, transcription was also measured from an rrnB P1 promoter derivative that did not contain any FIS sites. All promoters were cloned in pRLG770 and supercoiled DNA was used for the assays. The vector-encoded replication repressor, RNAI (∼110 nucleotides), is also indicated. (b) The repression afforded by FIS at each promoter is shown. The fold repression by FIS was calculated by dividing the activity in the absence of FIS by the activity in its presence. Values are the mean (with standard deviation) of three independent experiments. The activity of PguaB (-37 to +36) in the presence of FIS was too low to quantitate and hence a value for the fold repression was not obtained.