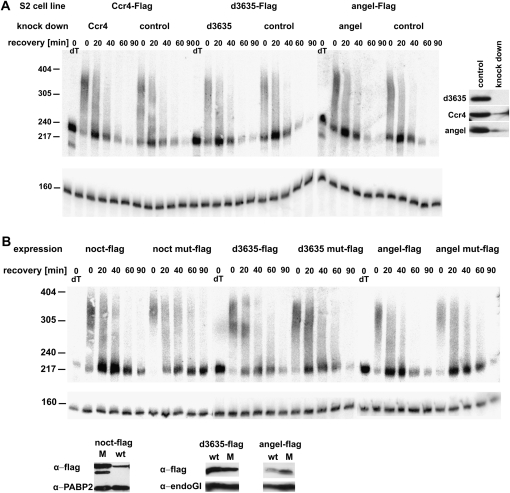
FIGURE 8.
Angel and 3635 do not play a detectable role in mRNA deadenylation (A) RNAi experiment: S2 cells stably transformed with expression constructs for Flag-tagged CCR4, Angel, or 3635 were treated with the corresponding dsRNA for 4 d under noninducing conditions. Then a 30-min heat shock at 35.6°C was applied. After return to 25°C, RNA was prepared at the indicated times of recovery and digested with RNase H in the presence of an hsp70-specific oligonucleotide. dT indicates Oligo(dT) was included in the RNase H digestion to mark the fully deadenylated RNA. Products were analyzed by Northern blot with probes against the 3′UTR of hsp70 and against U1 RNA serving as loading control. (Right panel) Expression of the flag-tagged proteins was induced for 4 h in small aliquots of the RNAi cells and in controls cells. Knock-down efficiency was examined by Western blot analysis with an anti-Flag antibody. Equal loading was controlled by Ponceau staining (data not shown). (B) Dominant-negative experiment: S2 cells expressing either Flag-tagged CCR4 paralogs or variants containing double point mutations in their exo III domains were grown under inducing conditions for 20 h, heat-shocked for 30 min at 35.5°C, and then allowed to recover at 25°C. RNA was prepared at the indicated times of recovery, and hsp70 mRNA was analyzed as above. RNA samples from nontransformed cells with and without addition of copper sulfate were analyzed in parallel and were indistinguishable from the samples expressing the wild-type CCR4 paralogs (data not shown). (Bottom panel) Expression of the tagged proteins was monitored by Western blotting with the antibodies indicated. EndoGI (Temme et al. 2009) and PABP2 served as loading controls.