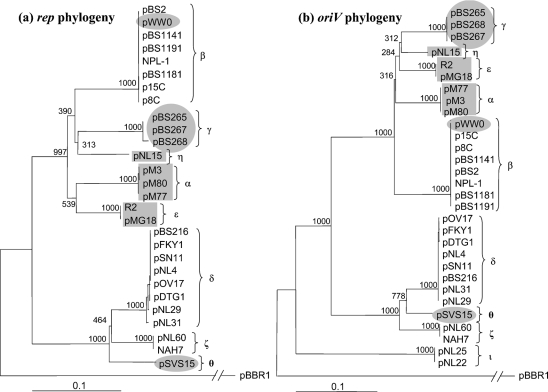
Fig. 1.
Neighbour-joining rooted phylogenies of the IncP-9 plasmid family based on sequence analysis of rep (a) and oriV (b) loci. Plasmid subgroups are bracketed and named with letters of the Greek alphabet from α to ι; grey background shapes define plasmid phenotypes: rectangle, multiple antibiotic resistance; oval, toluene/xylene degradation; circle, caprolactam degradation; no shape (clear background), naphthalene degradation. Bootstrap values (out of 1000 replicates) are shown adjacent to branch nodes. pBBR1 sequences were used to root the trees (pBBR1 branches are shortened for convenience). The lengths of horizontal branches correspond to evolutionary distances and the scale bars show the number of substitutions per site.