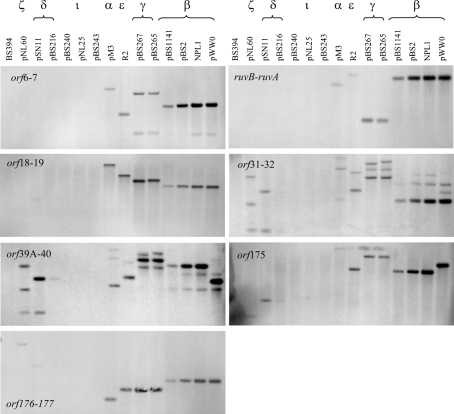
Fig. 3.
Hybridization of selected IncP-9 plasmids with pWW0-derived DIG-labelled probes specific to particular backbone loci. For hybridization, genomic DNA isolated from plasmid-containing bacteria was used; genomic DNA from plasmid-free P. putida BS 394 was used as a negative control (lane ‘BS394’). The DNA was digested with AvaI (orf6–7 and ruvB–ruvA loci) or SalI (others) before loading of the agarose gels, and was hybridized with pWW0-derived probes as described in Methods. In several hybridizations, multiple bands appeared in pWW0 and some test plasmids (orf6–orf7, orf31–orf32, orf39A–orf40). This might result from (i) incomplete digestion of the genomic DNA, (ii) non-specific hybridization or (iii) the presence of another region of homology elsewhere on the plasmid. The last could account for the appearance of multiple bands in the hybridizations of pWW0 and closely related β plasmids with orf31–32 and orf39A–40 probes, since pWW0orf32 and pWW0orf40 show 92.7 and 78.7 % nucleotide sequence identity to pWW0orf15 and pWW0orf171, respectively (Fig. 2).