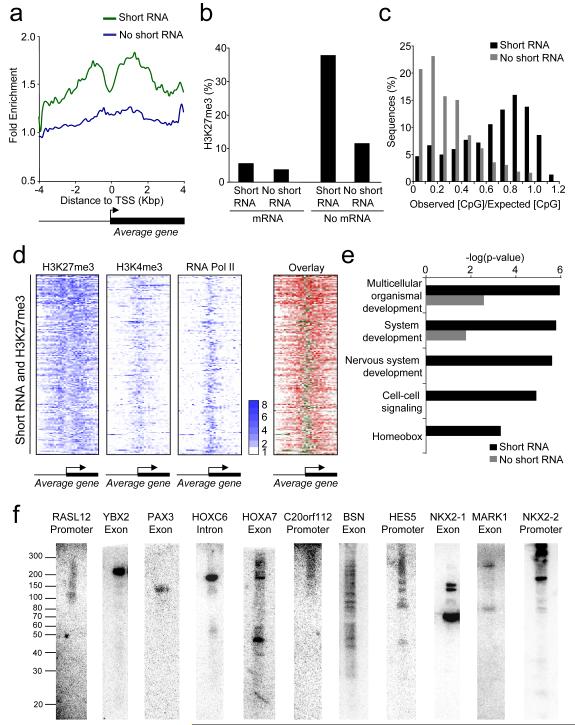
Figure 3. Short RNA loci are associated with H3K27me3.
A. Composite enrichment profile of H3K27me3 at genes for which no mRNA can be detected, divided into those that are associated with short RNA (green) and those not associated with short RNA (blue). The plot shows average fold-enrichment (normalized signal from H3K37me3 ChIP vs. total H3 ChIP).
B. Percentage of genes in different transcriptional categories that are associated with H3K27me3. Genes are first divided into those that produce detectable mRNA and those that do not and then into those that produce detectable short RNA and those that do not.
C. CpG content of short RNA loci not associated with mRNA. The data are plotted as a histogram of the observed CpG content divided by the expected CpG content and are compared to control genes that do not produce detectable short RNA or mRNA.
D. Heat maps showing enrichment of H3K27me3, H3K4me3 and RNA pol II at genes associated with short RNAs and H3K27me3. Each row represents one gene and each column represents the data from one probe, ordered by their position relative to the TSS. For the first three panels, fold enrichment is indicated by color, according to the scale on the right. The last panel overlays the RNA pol II (green) and H3K27me3 (red) to show their relative locations.
E. P-values for the enrichment of GO categories in the set of genes that are not associated with mRNA and transcribe short RNA (black bars) compared with those that do not produce detectable short RNA (grey bars).
F. Northern blotting for short RNAs transcribed from polycomb target genes in PBMC. The position of single-stranded RNA size markers are shown to the left. The genes and relative locations from which the short RNAs are transcribed are indicated above each blot.