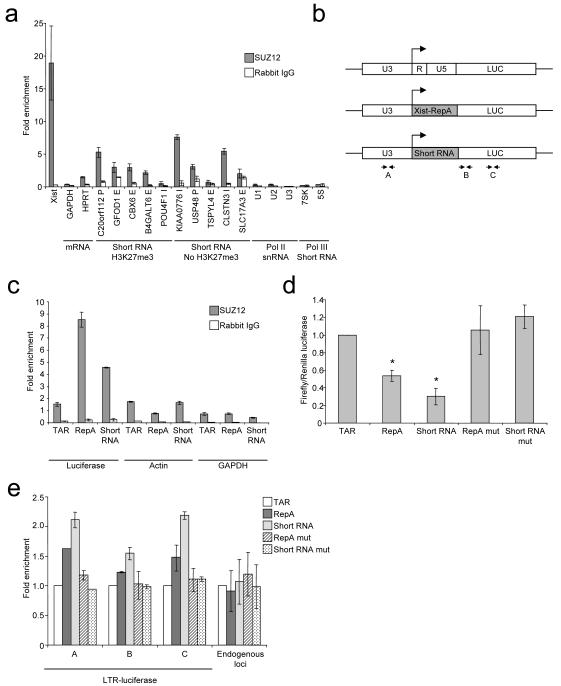
Figure 6. PRC2 interacts with short RNA in cells.
A. Fold enrichment (mean and SD, n=3) of different RNA species in SUZ12 IP (grey) and control IP (white) compared with input RNA and normalized to Actin measured by quantitative reverse-transcription PCR. The genes and relative locations from which the short RNAs are transcribed are indicated above (E=exon, I=intron, P=promoter).
B. Schematic showing the wild-type HIV LTR-luciferase construct and modified forms in which R and U5, encoding TAR, are replaced with the Xist-RepA stem-loop or the C20orf112 short RNA stem-loop. The position of primers used for RNA IP and ChIP experiments are marked.
C. Fold enrichment (mean and SD, n=3) of luciferase RNA containing different RNA stem-loops, actin RNA and GAPDH RNA in SUZ12 IP (grey) and control IP (white) compared with input RNA.
D. Luciferase activity in Hela cells transfected with plasmids encoding firefly luciferase downstream of wild-type or modified HIV LTRs. Firefly luciferase activity is plotted relative to co-transfected Renilla luciferase and normalized to wild-type LTR (mean and SD of 7 experiments (performed with 2 clones), each experiment comprising 3 measurements. * comparisons that gave significance at p<0.05.
E. Fold enrichment of H3K27me3 over H3 (mean and SD, n=3) across the different luciferase constructs (primers marked in B), normalized to the ratio measured at the wild-type LTR construct. Data for 3 endogenous loci (ACCN2, HAPLN2 and HMX2) are shown in comparison.