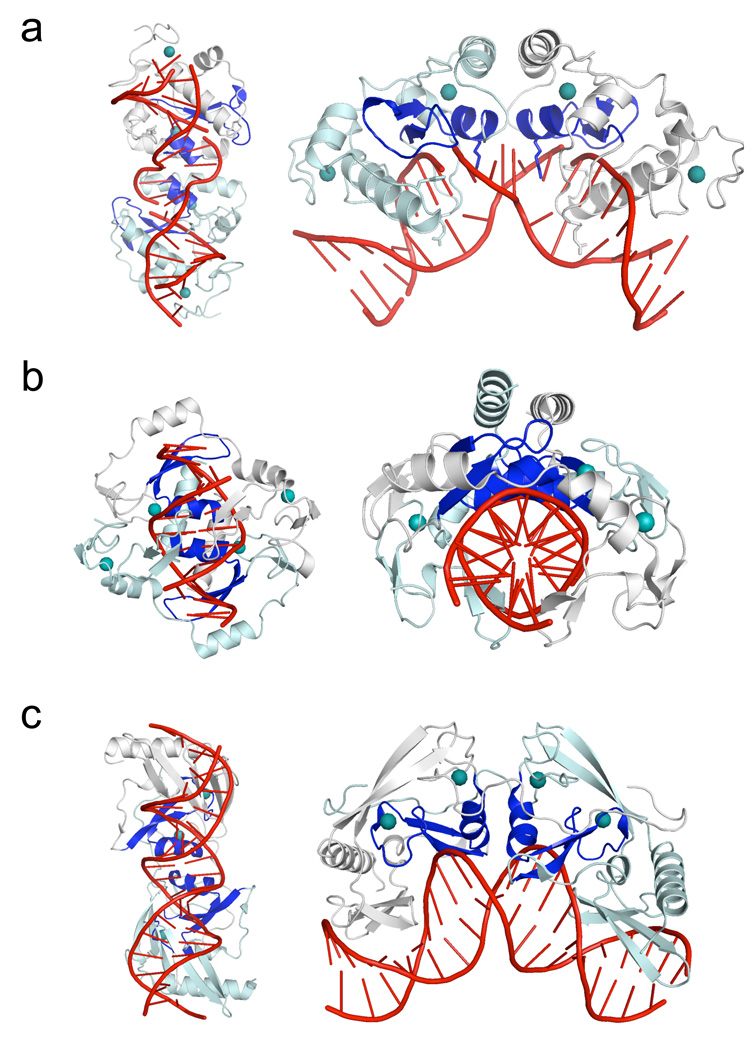
Figure 2. ββα-metal (HNH) endonucleases that contain two bound zinc ions per subunit.
For each endonuclease structure, the left panels show comparable orientations of the bound DNA targets, while the right panels show comparable orientations of the protein subunits (with the dyad symmetry axis running vertically and the protein subunits oriented to the left and right side of that axis). For all three structures, the ββα-metal catalytic motif is colored blue, and the bound zinc ions are shown as teal spheres. Panel a: The PacI restriction endonuclease. While the protein's overall organization of secondary structure elements, relative to the catalytic sites and bound zinc ions, is similar to Hpy99I, the overall architecture of the individual ββα-metal repeats, as well as the mode of DNA binding, is significantly different. Panel b: The core of the Hpy99I restriction endonuclease, which cleaves a five base pair CGWCG target, generating a 5-base, 3' overhang (Sokolowska et al., 2009). An N-terminal β-barrel domain (residues 1 to 53) that is not involved in DNA binding is removed for clarity. The remaining protein homodimer structure includes all catalytic and zinc-binding regions that correspond to those observed in PacI. Note that the orientation of the protein is roughly orthogonal to the major axis of the DNA duplex, which is relatively unbent. Panel c: The I-PpoI restriction endonuclease (a fourteen-base cutting homing endonuclease found in the eukaryotic amoeboid Physarum polycephalum) (Flick et al., 1998). The overall bend of the DNA target is similar to that displayed by PacI; however all base pairs are maintained in their original Watson-Crick base pairing arrangement. The protein fold, beyond the core ββα-metal motif, is completely different from that displayed by Hpy99I and PacI, indicating that divergence of these protein lineages may have occurred at an early stage, from a simple ββα-metal scaffold prior to subsequent elaboration and specialization. See also supplemental Figure S2.