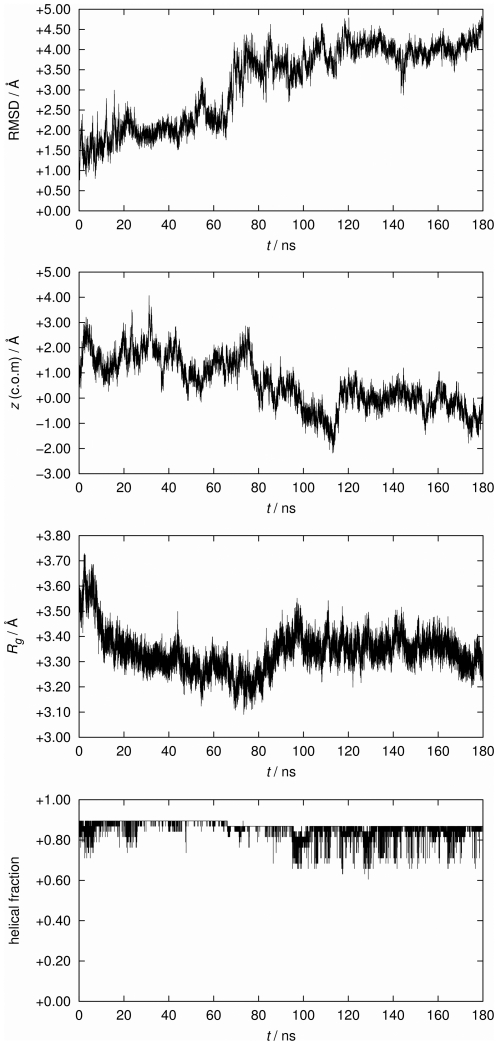
Figure 8. Dependence of various measures for protein stability over the simulation time after removal of the center-of-mass (c.o.m.) constraint (set to 0 ns).
From top to bottom: Root mean square deviation (RMSD) of the protein backbone, z coordinate (membrane normal) of the c.o.m. of the protein (corrected by removing the total membrane drift), the protein's radius of gyration (Rg), and the helical fraction recognized for the fold.