Fig. 1.
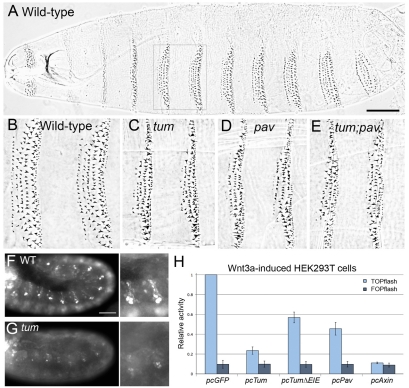
Tum and Pav control epidermal patterning in the Drosophila embryo and repress Wnt-induced TOPflash activity in HEK293T cells. (A) Wild-type embryos secrete a cuticle pattern with expanses of naked cuticle separating ventral denticle belts. (B) Higher magnification of ventral abdominal segments 2 and 3 indicated by box in A. Anterior is to the left in these and all subsequent panels. (C,D) Representative phenotypes of (C) tumAR2 and (D) pavB200 homozygous mutants show a reduced number of denticle rows in each belt compared with wild-type (B). (E) The most severe phenotypes observed among embryos segregating from the tum/+;pav/+ self-cross are not substantially different from either tum or pav single mutant phenotypes (see Table 1). Scale bar in A: 100 μm (A), 50 μm (for B-E). (F) rhomboid-lacZ marker is expressed in segmental stripes in the late epidermis. (G) tum mutant embryos show repression of this rho-lacZ expression. A GFP-marked balancer was used to distinguish homozygous mutants from their wild-type siblings (n>100 embryos at or later than stage 11). Note that rhomboid expression in the head (left in F and G) was detected at similar levels in both tum mutants and their wild-type siblings, providing an internal control for staining intensity. Scale bar: 50 μm (F,G); 25 μm (for zooms). (H) Tum represses TOPflash activation to 23% of pcGFP controls (P=0.001), when HEK293T cells are induced by Wnt-conditioned medium. Mutation of the GAP domain (pcTumΔEIE) does not eliminate the ability of Tum to repress TOPflash (P=0.008). Pav transfection represses activity to 45% of pcGFP controls (P=0.007). Axin, a known negative regulator of Wnt signaling, represses TOPflash activity to 11% of pcGFP (P<0.001). FOPflash values were not significantly altered by any of these treatments. Results are means ± s.d.