Figure 5.
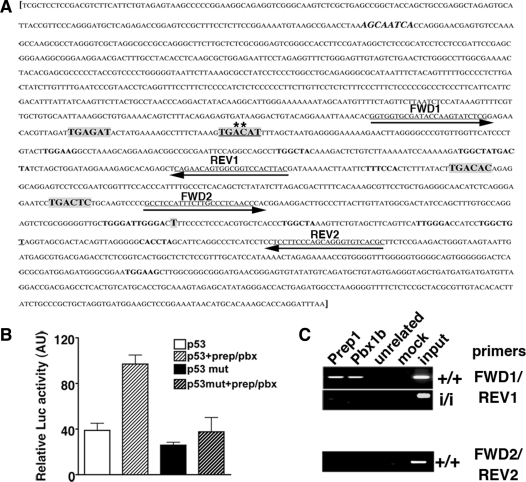
p53 expression is directly controlled by Prep1-Pbx complex. (A) Murine p53 gene regulatory region [M26862 (17)]. Putative Prep1-Meis/Pbx-binding sites are in bold and in a larger font (low-affinity sites are in bold with the same font as the rest of the sequence). The canonical sequence is underlined and mutated bases are marked with an asterisk (A to G and C to T). The sequence in bold and italic is a canonical binding site on the reverse strand. Sequences underlined and with arrows are the primers used for the PCR amplification of genomic DNA from ChIP (see below). Primer set 1 (FWD1 and REV1) spans the region containing the Prep1/Pbx canonical binding site. Primer set 2 (FWD2 and REV2) spans the control region not containing any Prep1/Pbx binding site. The transcription start site (T) is shown. (B) Luciferase activity, in arbitrary units, assayed from extracts of transiently transfected Cos7 cells. The reporter (luciferase) vector containing either the WT (p53); (17)) or mutated (p53mut) regulatory region (–1327/+337 bp) of the murine trp53 gene [whole sequence shown in (A)] was co-transfected with Prep1 and/or Pbx1b expression vectors. The graph shows the β-galactosidase-normalized luciferase activity of total cell extracts. (C) ChIP was performed as described in ‘Materials and Methods’ section. Cross-linked chromatin from Prep1+/+ and Prep1i/i MEF was immunoprecipitated with antibodies against Prep1, Pbx1b and uPAR (as unrelated antibody). Amplification was performed using a specific primer set for the trp53 regulatory or control region (containing or not the Prep1/Pbx1-binding sites, respectively) and PCR products were fractionated on a 2% agarose 1 × TAE gel and visualized by EtBr staining. The picture shows a representative experiment out of three performed with the same outcome. mut: mutated.