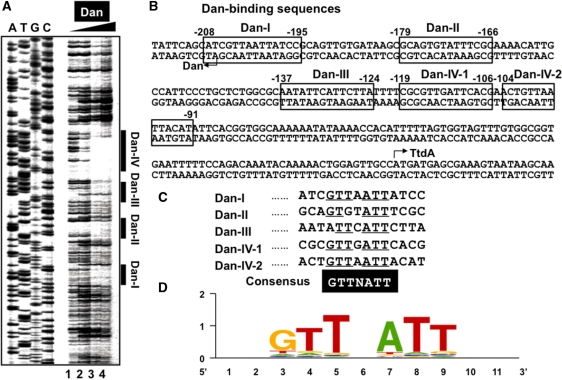
Figure 3.
Identification of the Dan box sequence. (A) Fluorescent-labeled SELEX segment (1.0 pmol) from the dan-ttdA spacer was incubated in the absence or presence of increasing concentrations of purified Dan (10, 20 and 40 pmol from left to right) and then subjected to DNase-I foot-printing assays. Lanes A, T, G and C represent the respective sequence ladders. (B) Four Dan-binding sites were identified on the dan-ttdA spacer sequence. (C) The Dan-box sequence GTTAAT was predicted after sequence comparison of four Dan-binding sites. Similar sequences were identified among a total of 688 Dan-binding fragments selected by Genomic SELEX. Using this Dan-box sequence, a total of about 1860 sites were predicted to be present in the entire E. coli genome. (D) Logo representation of Dan-binding site derived from sequences in in silico analysis. Logos were generated using weblogo (http://weblogo.berkeley.edu/).