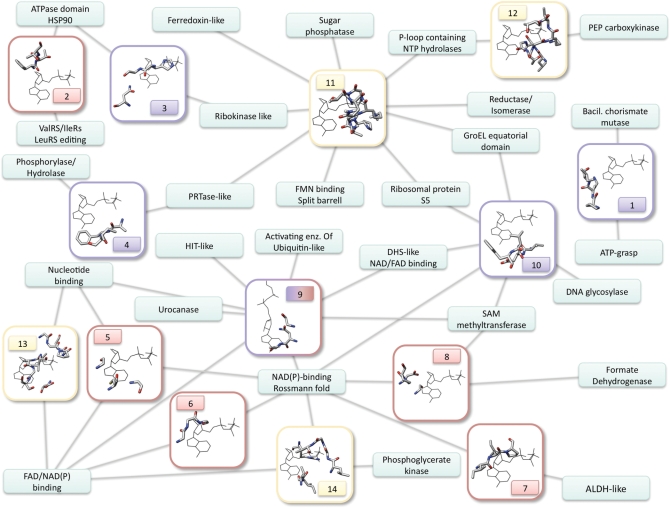
Figure 1.
Network representing the distribution of nucleotide-binding motifs across different protein folds. Each small rectangle is a fold, with the name written inside. The bigger rectangles represent structural motifs, numbered as in Supplementary Table S1, and with the frame and number colored according to the bound ligand fragment (lila: nitrogen base; red: ribose; yellow: phosphate). The edges connect each motif to the folds on which it was found. The network comprises 14 motifs and 27 folds. The pictures of the motifs show one representative structure belonging to one of the folds involved. All the ligands were aligned to a reference ligand by superimposing the relevant molecular fragments (i.e. for ribose-binding motifs, the ribose of the structure was aligned to the ribose of the reference ligand, etc.). The pictures, therefore, show the protein residues together with the reference ligand, which has always the same position. This was not possible for motif number 9, which binds both ribose and adenine in a conformation that cannot be aligned to the reference ligand. Accordingly, this is the only case in which the ligand in the picture is different.