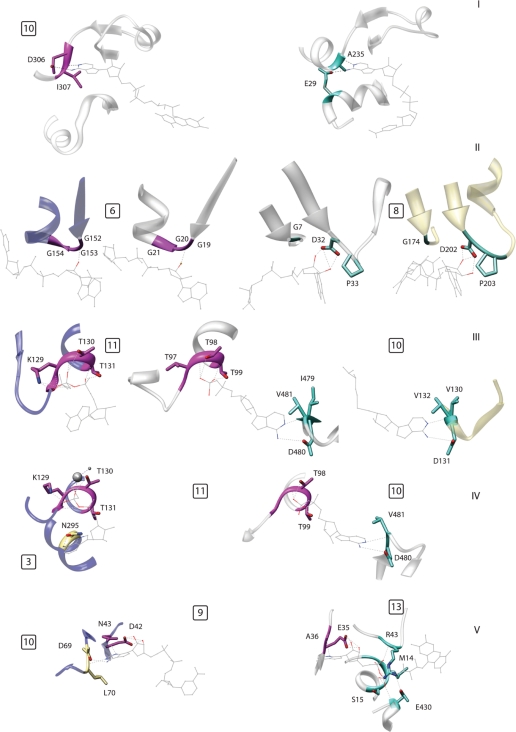
Figure 3.
Examples of modular composition of specific binding pockets; the numbers in the small squares indicate the motifs, numbered as in Figure 1 and Supplementary Table S1. (I) Two proteins belonging to the same fold (DHS-like NAD/FAD-binding domain) that bind the same ligand fragment, namely adenine, in different ways. Left: ETF from M. methylotrophus (PDB: 1o96); right: lysine deacetylase of the Sir2 family (1s7g). (II) An example of alternative motifs in a single fold (Rossmann fold). The two central proteins are the Siroheme synthase CysG from Salmonella typhimurium (1pjs, inner left) and glyceraldehyde-3-phosphate dehydrogenase (1ihx, inner right). These proteins belong to the same fold but use different ribose-binding motifs shared with another fold. Outer left: thioredoxin reductase (1f6m, FAD/NAD(P)-binding domain), outer right: formate dehydrogenase H (1fdi, formate dehydrogenase/Dimethyl Sulfoxide reductase fold). (III) The center protein shares a phosphate binding motif with the one on the left and an adenine motif with the one on the right. All three proteins belong to different folds. Left: MurC (1gqy, ribokinase-like fold); center: group II chaperonin from Thermococcus strain KS-1 (1q3s, GroEL equatorial domain-like); right: methyltransferase from Dengue virus (1l9k, SAM-dependent methyltransferases). (IV) Two proteins sharing a phosphate-binding motif but using two different motifs to bind adenine. Left: MurC (1gqy, ribokinase-like fold); right: group II chaperonin from Thermococcus (1q3s, GroEL equatorial domain like). (V) The same motif, binding the sugar edge of adenine, is associated with a motif binding the ‘Watson–Crick’ edge (left) and with a phosphate-binding motif (right) in two different structures. Left: Gal10p (1z45, NAD(P)-binding Rossmann fold); right: polyamine oxidase (1b37, FAD/NAD(P)-binding domain).