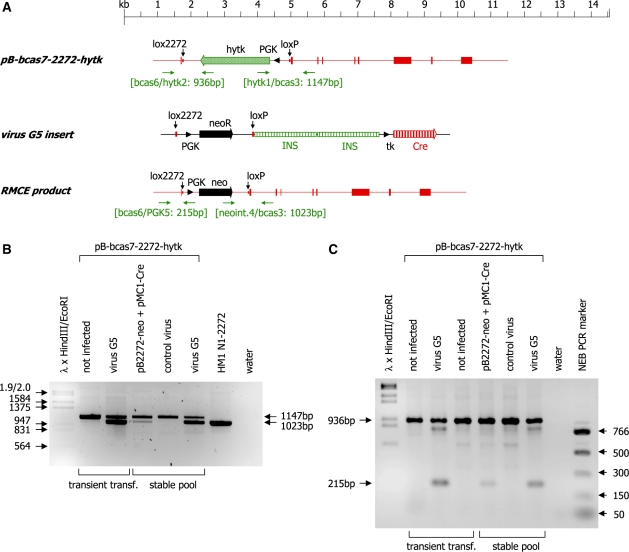
Figure 7.
(A) Schematic representation of the plasmid pB-bcas7-2272-hytk, the insert of the virus vector G5 and the resulting RMCE product. Exons of the β-casein gene are indicated as shaded boxes, the neomycin and hytk selection marker genes are indicated as solid and shaded arrows, respectively and the Cre ORF (Cre) is indicated as a vertically striped arrow. The PGK promoter elements directing expression of the selection marker genes are indicated as black arrowheads. The positions of the lox2272 and loxP sites are marked by vertical arrows. The primer binding sites (horizontal arrows) used for genotyping and the sizes of the expected PCR products are indicated. (B) PCR analysis of DNA isolated from HEK 293 cells transiently or stably transfected with pB-bcas7-2272-hytk. The cells were either co-transfected with the indicated plasmids or infected with the indicated virus vectors 24 h post-transfection. The 3′ end of the RMCE reaction was analysed using the primer combination bcas3/neoint.4/hytk1. Successful recombination at the loxP site is indicated by the presence of the 1023-bp PCR product. Non-recombined DNA yields a PCR product of 1147 bp. Phage λ DNA digested with HindIII and EcoRI was used as molecular weight marker. (C) The 5′ end of the RMCE reaction was analysed using the primer combination bcas6/PGK5/hytk2. Successful recombination at the lox2272 site is indicated by the presence of the 215-bp PCR product. Non-recombined DNA yields a PCR product of 936 bp. Phage λ DNA digested with HindIII and EcoRI and the NEB PCR marker (New England Biolabs) were used as molecular weight markers.