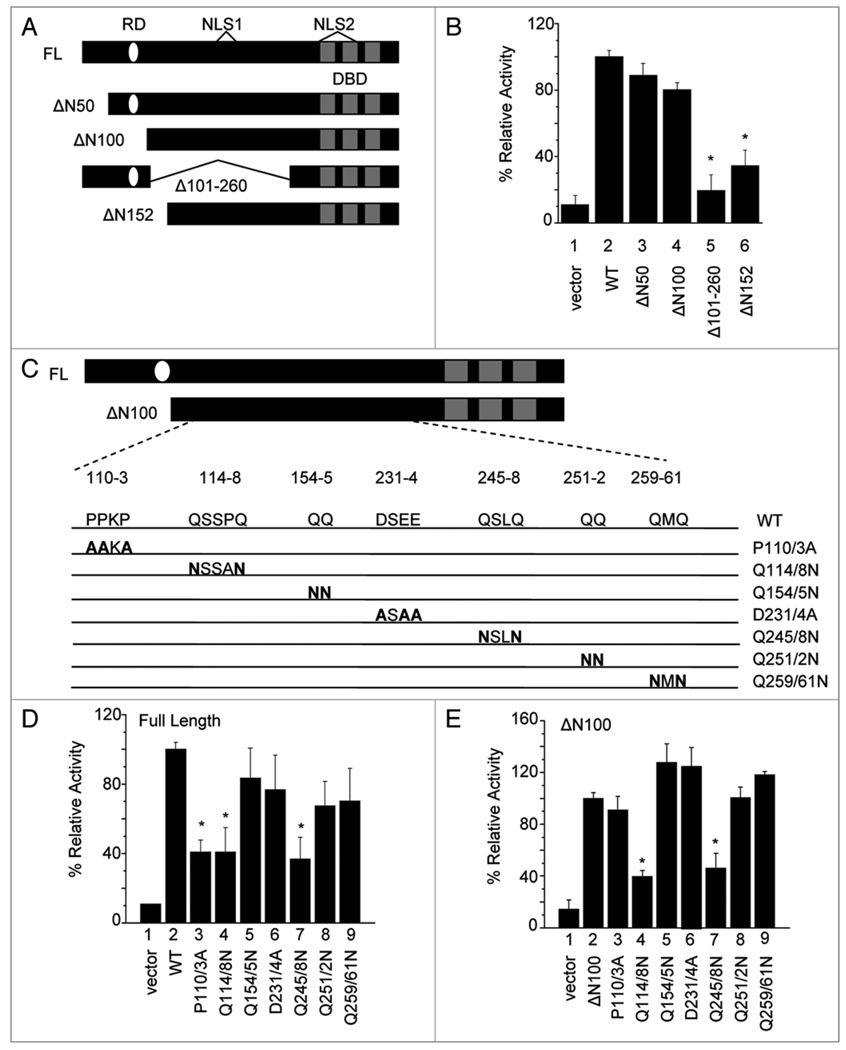
Figure 1.
Mutation of regions Q114-Q118 and Q245–248 causes a significant decrease in cyclin D1 promoter activity. (A) Schematic diagram of KLF8 deletion mutants. The three zinc finger DNA binding domain (DBD), two nuclear localization signals (NLS) and the repression domain (RD) are depicted. (B) Luciferase activity was measured in NIH3T3 cells co-transfected with the cyclin D1 promoter reporter and the KLF8 deletion mutants. The empty vector and wild type KLF8 (WT) or a KLF8 construct lacking its repression domain (ΔN100) were used as negative and positive controls, respectively. After 16 h, cells were harvested for luciferase assays as described in Materials and Methods. Results were normalized to WT. (C) Schematic diagram of point mutations generated in the context of either full length KLF8 (FL) or ΔN100. Prolines (P) or aspartates (D) were mutated to alanines (A) and glutamines (Q) were mutated to asparagines (N). (D and E) NIH3T3 cells were co-transfected with the cyclin D1 promoter reporter and the point mutants in the context of either full length (D) or ΔN100 (E). Luciferase assays were performed similarly as described in (B). Results were normalized to either WT (D) or ΔN100 (E). Equal expression of the KLF8 constructs is shown in Supplemental Figure 1. All experiments were done in triplicate and the results represent mean ± S.E. of at least three experiments. *p < 0.05 compared to WT (B and D) or ΔN100 (E).