Abstract
The appearance of oxygen in the Earth's atmosphere via oxygenic photosynthesis required strict anaerobes and obligate phototrophs to cope with the presence of this toxic molecule. Here we show that in the anoxygenic phototroph Rubrivivax gelatinosus, the terminal oxidases (cbb3, bd, and caa3) expand the range of ambient oxygen tensions under which the organism can initiate photosynthesis. Unlike the wild type, the cbb3−/bd− double mutant can start photosynthesis only in deoxygenated medium or when oxygen is removed, either by sparging cultures with nitrogen or by co-inoculation with strict aerobes bacteria. In oxygenated environments, this mutant survives nonphotosynthetically until the O2 tension is reduced. The cbb3 and bd oxidases are therefore required not only for respiration but also for reduction of the environmental O2 pressure prior to anaerobic photosynthesis. Suppressor mutations that restore respiration simultaneously restore photosynthesis in nondeoxygenated medium. Furthermore, induction of photosystem in the cbb3− mutant led to a highly unstable strain. These results demonstrate that photosynthetic metabolism in environments exposed to oxygen is critically dependent on the O2-detoxifying action of terminal oxidases.
Keywords: Cytochrome Oxidase, Gene Regulation, Oxidative Stress, Oxygen Toxicity, Photosynthesis, Anaerobic Metabolism, Oxygen Regulation
Introduction
Primary production of oxygen on Earth, accomplished by oxygenic photosynthetic organisms (1, 2), placed strict anaerobes in a critical situation. They had to deal with the continuous presence of O2 to survive, and the redox situation was probably further complicated for the phototrophs because of their reactive photosynthetic metabolism. A way to overcome the presence of oxygen is to reduce the O2 pressure, mediated by oxygen-consuming enzymes of facultative anaerobes. Obligate anaerobes had to acquire or replace these anaerobic enzymes with oxygen-dependent versions, building O2-dependent metabolic networks (3, 4). Otherwise, they would have to depend on O2-consuming organisms to sustain their growth and expansion.
The effect of O2 on nitrogen fixation is one of the best examples of how bacteria dealt with O2. In diazotrophic cyanobacteria, oxygen evolving photosynthesis is in conflict with the nitrogen fixation process. Because the nitrogenase, an enzyme catalyzing the reduction of nitrogen to ammonium, is extremely sensitive to oxygen, several strategies have evolved to protect this enzyme from inactivation by O2. Many filamentous cyanobacteria develop heterocysts to exclude O2. Tomitani et al. (5) suggest that the evolution of heterocysts may have accompanied the appearance of oxygen on Earth. In Anabaena sp. PCC7120, a decrease of photosystem II activity and an increase of aa3 cytochrome c oxidase activity in the heterocysts has been shown (6, 7). For some nonheterocystous cyanobacteria, N2 fixation takes place in the dark to avoid production of O2 by photosynthesis (8). In aerobic and microaerophilic diazotrophic bacteria, terminal oxidases including cbb3 and bd oxidases have been shown to be important for N2 fixation. For instance, in the endosymbionts Bradyrhizobium japonicum and Rhizobium meliloti, cbb3 cytochrome c oxidase is required to fix N2 in symbiosis (9). Furthermore, in Azotobacter vinelandii, the bd quinol oxidase null mutant is unable to fix nitrogen (10). Many strict anaerobes of the Bacteroides class of eubacteria cannot grow in the presence of micromolar dissolved oxygen. However, they can survive in oxygenated environments until the O2 pressure is reduced by facultative anaerobes. Bacteroides fragilis encodes a cytochrome bd oxidase that is essential for O2 consumption (11). The increasing amount of data from bacterial genomes reveals the presence of genes encoding terminal oxidases not only in aerobes but also in strict anaerobes. The occurrence of terminal oxidases in anaerobic microorganisms remains intriguing and suggests that these complexes may have roles other than respiration in these organisms.
The main challenge for phototrophs is to reconcile O2 and light. The harmful effects of O2 and the excess of light energy arise from the production of bacteriochlorophyll triplet states. In aerobic conditions, the triplet-excited states of bacteriochlorophylls sensitize oxygen to produce singlet molecular oxygen. In the photosystem, carotenoids bound in proximity to bacteriochlorophylls prevent formation of this reactive oxygen (12). In addition to this scavenger, photosynthetic bacteria (Rhodobacter sphaeroides and Rhodobacter capsulatus) have established tight redox regulation mechanisms (13, 14) that serve to resolve this incongruity between light and atmospheric or medium dissolved O2. In these bacteria, the cbb3 cytochrome c oxidase is also involved in the regulation of photosynthesis gene expression. In R. sphaeroides, it was suggested that under aerobic conditions, cbb3 oxidase acts as an O2 sensor that generates an inhibitory signal to repress photosynthesis gene expression via the two-component system PrrBA (RegBA). In R. capsulatus, it was proposed that the quinol pool redox state rather than cbb3 cytochrome c oxidase regulates photosynthesis gene expression via the RegBA two-component system.
This study focuses on the phototrophic Rubrivivax gelatinosus terminal oxidases and their requirement for photosynthetic and aerobic growth. We have previously detailed the AcsF/BchE enzyme replacement and suggested a similar HemF to HemN replacement during the shift from aerobic to anaerobic environment to emphasize the O2-dependent and -independent metabolic networks (15, 16). Here, we further investigate the problem of O2 and photosynthesis (PS)2 in this bacterium and show how the terminal oxidases expand the physiological range of O2 environments under which R. gelatinosus and probably many other facultative phototrophs can carry out photosynthesis.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Media
Escherichia coli was grown at 37 °C on LB medium. R. gelatinosus was grown at 30 °C, in the dark, aerobically (high oxygenation, 250-ml flasks containing 20 ml of medium), semi-aerobically (250-ml flasks containing 120 ml of medium), microaerobically (low oxygenation, 50-ml flasks filled with 50 ml of medium), or in light (photosynthesis) in filled and sealed tubes in nondeoxygenated malate growth medium (15). Antibiotics were used at the following concentrations: kanamycin (Km), 50 μg/ml; ampicillin, 50 μg/ml; streptomycin, 50 μg/ml; spectinomycin, 50 μg/ml; tetracycline, 3 μg/ml; and erythromycin, 10 μg/ml. The bacterial strains and plasmids are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strains or plasmids | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-blue | supE44, hsdR17, recA1, endA1, gyrA46, thi-1, relA1, lac−F'[proAB+, lacI, lacZΔM15 Tn10(Tcr)] | Stratagene |
| R. gelatinosus | ||
| Strain S1 | Wild type | Ref. 33 |
| cbb3− | ccoN strain (ccoN::Km) | This work |
| bd− | cydA strain (bd::Ω) | This work |
| bo− | cyoA strain (bo::Km) | This work |
| caa3− | coxBA strain (coxBA::Km) | This work |
| cbb3−bd− | ccoN::Km, cydA::Ω | This work |
| ppsRK | ppsR::Km | Ref. 20 |
| cbb3−bd− | ppsR−ppsR::Km, ccoN::Ery, cydA::Ω | This work |
| cbb3−ppsR− | ccoP::Ω, ppsR::Km | This work |
| bd−ppsR− | cydA::Ω, ppsR::Km | This work |
| Plasmids | ||
| pGEMT | Cloning vector (Apr) | Stratagene |
| pUC4K | Plasmid bearing the Km cartridge (AprKmr) | Pharmacia |
| pDW9 | Plasmid bearing the Ω cartridge (Apr spr Smr) | Ref. 34 |
| pBBR1MCS-3 | Expression vector with tetracycline resistance | Ref. 35 |
| pA200 | ppsR::Km | Ref. 20 |
| pA410 | pBBR1MCS-3 bearing ppsR gene | Ref. 20 |
| pBS9 | pGEMT+ 1.7-kb fragment containing coxBA | This work |
| pBS10 | pGEMT+ 1.9-kb fragment containing ccoN | This work |
| pBS15 | pGEMT+ 1.6-kb fragment containing cydA | This work |
| pBS16 | pGEMT+ 2.3-kb fragment containing cyoA | This work |
| pBS19 | Km cartridge cloned into NarI site within coxBA from pBS9 | This work |
| pBS20 | Km cartridge cloned into MscI site within ccoN from pBS10 | This work |
| pBS22 | Erythromycin cartridge cloned into MscI site within ccoN from pBS10 | This work |
| pBS28 | Ω cartridge (BamHI) cloned into site within ccoP (BamHI) | This work |
| pBS25 | Ω cartridge cloned into XmaI site within cydA from pBS15 | This work |
| pBS26 | Km cartridge cloned into BglII site within cyoA from pBS1–6 | This work |
| pBS27 | fnrL gene (SnaBI-ApaI) cloned in pBBR1MCS-3 (SmaI-ApaI) | This work |
| pBS32 | ccoP gene (SacII) cloned in pBBR1MCS-3 (SacII) | This work |
Membrane Preparation and Pigment Extraction
The membranes and pigments were prepared as described previously (15).
Molecular Biology Techniques
Standard methods were performed according to Sambrook et al. (17) unless indicated otherwise. The sequences reported in this paper were deposited in GenBankTM with accession numbers AY648960 (ccoNOPQ), AY655709 (cydBA), and AY648055 (coxBAC)
Gene Cloning and Plasmid Constructions for Allele Replacement
To clone the ccoN gene from genomic DNA of R. gelatinosus, a 1.9-kb fragment was amplified by PCR using the primers ol-326 and ol-302. The fragment was cloned into the PCR cloning vector pGEM-T to give pBS10. The ccoN gene was inactivated by the insertion of the Km or erythromycin cassette at the unique MscI site within the ccoN coding sequence. Briefly, plasmid pBS10 was subjected to restriction enzyme digestion and treated with Klenow polymerase, prior to ligation with the 1.2-kb EcoRI blunt digested Km cassette conferring resistance to kanamycin or SmaI-digested erythromycin cassette (0.9 kb). The resulting recombinant plasmids were designated pBS20 and pBS22. The primers used in this work are listed in Table 2.
TABLE 2.
Primers
| Gene | Primer | Sequence (5′ to 3′) |
|---|---|---|
| ccoN | 326 | CCTGTACCTGCTCATCCC |
| 302 | GTGGCCGTAGCGCAGCG | |
| cydA | 367 | CGTCGGCGGCCTGGCAGC |
| 369 Rev | GTCGGGGTCTGCACGTCG | |
| cyoA | bo For | ACGCACCGGGGCGCTACC |
| bo Rev | CAGCAGGTGCCCGGCCCC | |
| coxBA | B21 | GGGTGATGTTCTGGTCCA |
| B12 | GAAGTGGCGGTCGGTCAG |
A 1.7-kb coxBA fragment was cloned into pGEM-T using the primers B21-B12 to give pBS9. coxB gene was inactivated by the insertion of the Km cassette at the unique NarI site within the coxB coding sequence. To that aim, pBS9 was subjected to restriction enzyme digestion and treated with Klenow polymerase, prior to ligation with the 1.2-kb EcoRI blunt digested Km. The resulting recombinant plasmid was designated pBS19.
To clone the cydA gene from genomic DNA of R. gelatinosus, a 1.6-kb fragment was amplified by PCR using the primers 367 and 369 Rev. The fragment was cloned into pGEMT to give pBS15. The cydA gene was inactivated by insertion of the Ω cassette in the unique XmaI site. Plasmid pBS15 was subjected to digestion with XmaI, prior to ligation with the 2-kb XmaI-digested Ω cassette that confers resistance to spectinomycin and streptomycin. The resulting recombinant plasmid was designated pBS25.
To clone the cyoA gene from genomic DNA of R. gelatinosus, a 2.3-kb fragment was amplified by PCR using the primers bo For and bo Rev. The fragment was cloned into pGEM-T to give pBS16. The cyoA gene was inactivated by the insertion of the Km cassette in the unique BglII site within the cyoA. Plasmid pBS16 was subjected to restriction enzyme digestion BglII, prior to ligation with the 1.2-kb BamHI-digested Km cassette. The resulting recombinant plasmid was designated pBS26.
Gene Transfer and Strain Selection
Transformation of R. gelatinosus cells was carried out by electroporation as described previously (15). Transformants were selected on malate plates supplemented with the appropriate antibiotic either under aerobic conditions or anaerobic photosynthesis in sealed bags with H2- and CO2-producing gas packs (Anaerocult; Merck). Subsequently, following transformant selection, template genomic DNA was prepared from the transformants, and confirmation of the presence of the antibiotic resistance marker at the desired locus was performed by PCR.
Blue Native Gel Electrophoresis
Blue native (BN)-polyacrylamide gel electrophoresis and in-gel cytochrome c oxidase catalytic activity assays were performed as described in Refs. 18 and 19. 2 mg of membrane proteins were solubilized for 30 min on ice in 0.1 m Tris buffer containing 1% N-dodecyl β-d-maltoside, 10% glycerol, and 0.3 m NaCl. The supernatant was cleared by centrifugation at 70,000 rpm for 15 min. 4.5 μl of load buffer (5% blue G250 and 500 mm n-aminocaproic acid) were added to the supernatant. BN-PAGE electrophoresis was performed with few modifications; the gel buffer contained 75 mm Bis-Tris and 0.75 m n-aminocaproic acid, pH 7.0, and in the cathode buffer, 0.03% of N-dodecyl β-d-maltoside were added. The cytochrome c oxidase activity was revealed with the 3,3′-diaminobenzidine tetrahydrochloride. Acid-labile hemes were extracted from washed membranes and analyzed by HPLC on a reverse phase C18 column with a acetonitrile gradient in 0.1% trifluoroacetic acid/water.
NADI and TMPD Reactions
The cytochrome c oxidase activity was detected by staining the colonies with a 1:1 mixture of 35 mm α-naphtol dissolved in ethanol and 30 mm NADI. The colonies that exhibit cytochrome c oxidase activity turn blue after the addition of substrate.
The artificial electron donor TMPD can be oxidized by the cytochrome c oxidase to form a blue indophenol compound. Cytochrome c oxidase activity was measured spectrophotometrically on membranes in 0.1 m Tris buffer, pH 7, by following the increase in the A562 nm after the addition of 0.5 mm TMPD at room temperature (carry 500 spectrophotometer).
Oxygen Consumption Assay
To monitor the oxygen tension in the culture medium, the cells were removed by centrifugation, and the oxygen concentration was measured in the medium with a Clark-type oxygen electrode.
RNA Extraction and Real Time RT-PCR
Total RNA from wild type and cbb3− cells grown photosynthetically or microaerobically was extracted according to Ref. 20. The RNA concentration was determined by absorption at 260 nm. Specific primers were designed to amplify (200 nucleotides) fragment of R. gelatinosus pufC, bchE, and bchC genes. The primers used for RT-PCR and qPCR were: bchE reverse (5′-cttggcgatctcgaagttgt-3′), bchE forward (5′-ctgaaggcgttcgtgaagag-3′), bchC reverse (5′-ccagtcgaggaccatcttc-3′), bchC forward (5′-gccgctgcacttcaatttc-3′), pufC reverse (5′-gcggcttccacggcagaggcg-3′), and pufC forward (5′-gcggcttccacggcagaggcg-3′). Reverse primers for each gene were annealed to 2 μg of total RNA extracted and extended for 50 min in the presence of 1 mm dNTPs at 55 °C using 200 units of reverse transcriptase (Superscript III; Invitrogen). 2 μl of the reverse transcriptase reaction was used for the subsequent PCR; each PCR contained 50 units of Taq DNA polymerase (Qiagen GmbH, Hilden, Germany) and 5% dimethyl sulfoxide. The PCR program included one cycle of 95 °C for 1 min, 30 cycles of 94 °C for 10 s, 54 °C for 30 s, 72 °C for 30 s, and a final incubation at 72 °C for 10 min. Amplified products were analyzed by electrophoresis in a 1.2% agarose gel.
qPCR
Levels of specific transcripts in total mRNA were quantified by qPCR using the Engine Light cycler 480 from Roche Applied Science. Single-stranded cDNA, synthesized in an reverse transcriptase reaction using DNase-treated RNA (as described above), served as the template for qPCR amplifications, which were performed using the Light cycler 480 SYBR GreenI Master qPCR Kit (Roche Applied Science). The specific amplification protocol was 1 cycle at 95 °C for 10 min; 44 cycles at 94 °C for 10 s, 56 °C for 15 s, 72 °C for 8 s; and a final incubation of 72 °C for 10 min. We determined the absolute level of each specific RNA (21). The relative expression (or the n-fold change) of the target genes was calculated by comparing the RNA in the cbb3− with the wild type.
RESULTS AND DISCUSSION
The Aerobic Respiratory Chain of R. gelatinosus
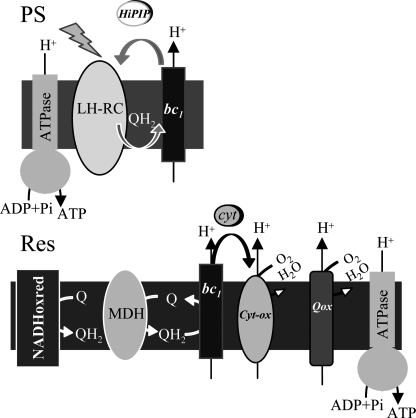
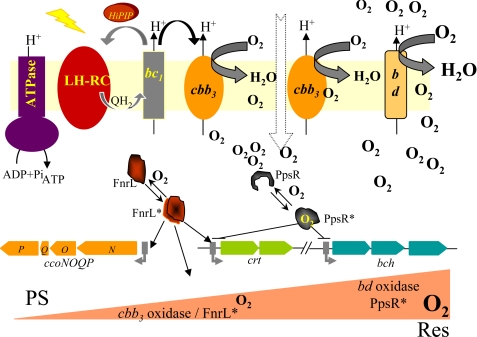
R. gelatinosus is a facultative photosynthetic bacterium that requires oxygen as exogenous electron acceptor during aerobic respiration. Genetics together with sequence analyses showed that its respiratory electron transport is branched after the quinone pool into one or several bc1-independent pathways on one side and an electron transfer chain coupling the bc1 complex to cytochrome c oxidases on the other side (22) (Fig. 1). PCR-based cloning allowed the identification of four oxidase encoding operons (supplemental Fig. S1). Two operons ccoNOQP and coxBAC code for the putative cbb3 and caa3 cytochrome c oxidases, respectively, and two other operons cydBA and cyoBAC are coding for putative bd and bo quinol oxidases. Single mutations were generated to disrupt each operon with antibiotic cassettes, and growth of the mutants was followed under photosynthesis and different aerobic conditions (supplemental Fig. S2).
FIGURE 1.
The electron transfer chain in the R. gelatinosus membrane. QH2 hydroquinone production occurs in the reaction center (RC) under photosynthesis and in the dehydrogenases (NADHoxred and malate dehydrogenase (MDH)) complexes under respiration. Under photosynthesis, the reaction center is reduced by the cytochrome bc1 complex via the HiPIP ferredoxin. Quinol oxidases (Qox) use QH2 to reduce oxygen to water. Cytochrome oxidases (Cyt-ox) are reduced by the cytochrome bc1 complex via the soluble cytochrome c.
The cbb3− mutant showed a slow respiratory growth; this phenotype was more pronounced under microaerobic conditions, suggesting that this oxidase is more required under these conditions (supplemental Table S1). In agreement with this, the expression profile of cbb3 revealed that this oxidase is highly induced under microaerobiosis.3 Furthermore, this cbb3− mutant is NADI− and TMPD−, indicating that cbb3 is the only active cytochrome c oxidase in the wild type strain. Disruption of bd oxidase also resulted in a respiration growth defect, especially under aerobic conditions. Under microaerobic conditions the bd− mutant doubling time is comparable with that of wild type, indicating that the bd oxidase is more required under high aeration. (A summary of the growth phenotypes of the mutants is presented in supplemental Table S1). In contrast, inactivation of caa3 and bo oxidases presented no difference of phenotype compared with the wild type. From these data the presence of two terminal oxidases in wild type R. gelatinosus is inferred: (i) a cbb3 cytochrome c oxidase induced under microaerobic conditions and which receives electrons from the bc1 complex and (ii) a bd type quinol oxidase required for aerobic conditions.
Photosystem Synthesis Requires the Terminal Oxidases under Aerobic Conditions
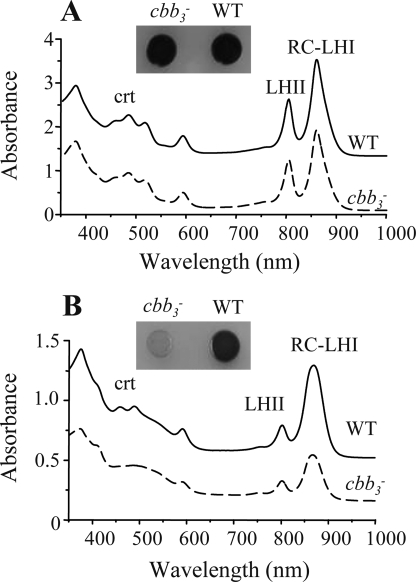
The R. gelatinosus cytochrome bc1− mutant was pale and less pigmented than the wild type when grown aerobically (22). This suggested that the bc1-dependent respiratory branch is involved in the synthesis of pigments and pigment-binding proteins. Similar to the bc1− mutant, the aerobically grown cbb3−- and bd−-deficient single mutant cells were also pale compared with the wild type (supplemental Fig. S2). To assess the effect of this mutation on the synthesis of bacteriochlorophyll a- and bacteriochlorophyll-binding proteins (RC-LHI/LHII), the wild type and the cbb3− mutant were grown under semi-aerobic or photosynthesis conditions, and equal amounts of membranes prepared from these cells were analyzed by spectroscopy. As shown in Fig. 2, spectral analyses of the membranes (Fig. 2A) and extracted pigments (supplemental Fig. S3) showed that both wild type and cbb3− mutant produced comparable amounts of photosynthesis complexes under photosynthesis conditions. Under semi-aerobic conditions, a substantial reduction in the absorbance at 800–870 nm was observed in the cbb3− mutant compared with the wild type, indicating a significant reduction of the level of the LHII and RC-LHI complexes (Fig. 2B). Spectral analysis of extracted pigments revealed that the cbb3− mutant produced lower amounts of bacteriochlorophyll a as judged by the peak at 770 nm (supplemental Fig. S3). These results indicate that full bacteriochlorophyll and photosynthesis complex production under oxygenated conditions requires the presence of the cbb3 cytochrome c oxidase in membranes.
FIGURE 2.
Absorption spectra of membranes from wild type (WT, solid line) and cbb3− mutant (dashed line) cells grown under photosynthesis (A) and semi-aerobis conditions (B). RC and LHI/II are the reaction center and the light-harvesting complexes, respectively. The less pigmented phenotypes of the cbb3− mutant under respiratory conditions is also seen when cells are growing on solid medium unlike cells growing under photosynthesis (supplemental Fig. S3). The zero level of the upper spectra was shifted for better viewing.
Furthermore, following inoculation from a semi-aerobic culture into photosynthesis-permitting conditions, the wild type strain resumed growth after a lag phase of 4 h (supplemental Fig. S4), whereas the cbb3− strain presented a lag phase of 7 h. Once photosynthetic growth started, the doubling times were comparable (2.5 h for both). The difference in growth following the shift from semi-aerobic to photosynthetic growth is probably due to the lower amount of photosystem in the cbb3− mutant under semi-aerobic conditions and also to low photosynthetic gene transcription in the cbb3− mutant when inoculated into the fresh nondeoxygenated medium. Because it is well established that the expression of these genes is repressed by oxygen in phototrophic bacteria including in R. gelatinosus, the transition of the wild type and the cbb3 mutant to photosynthesis should require de novo synthesis of photosynthetic complexes to support photosynthetic growth. However, the presence of O2 in the medium will inhibit photosynthetic gene expression until the oxygen is depleted from the medium. One may anticipate then that inactivation of one or more components of the respiratory electron transport chain would affect photosynthetic gene expression. A similar but less pronounced effect was obtained with the bd− mutant (supplemental Fig. S2). This mutant was also less pigmented than the wild type. The difference in response to oxygen between the cbb3− and bd− mutants is due to the fact that cbb3 is highly induced and more active under semi and microaerobiosis. These data show that terminal oxidases are necessary for efficient production of photosystem components.
To further demonstrate the importance of cbb3 activity in the induction of photosynthesis gene expression in R. gelatinosus, we monitored the accumulation of pufC, bchE, and bchC transcripts in the cbb3− mutant and the wild type grown under photosynthesis and microaerobic conditions. RT-PCR and qPCR showed a significant decrease of these transcripts in the cbb3− mutant grown under microaerobic conditions (supplemental Fig. S5). The transcripts of pufC and bchC were 10-fold less abundant in the mutant compared with wild type. A 26-fold decrease of bchE transcript was recorded in the cbb3− mutant. No significant difference in the level of these transcripts could be detected when cells were grown photosynthetically. These data show the requirement of cbb3 for the full expression of these photosynthesis genes.
cbb3 and bd Oxidases Are Required for Photosynthetic Growth at Low O2 Concentrations
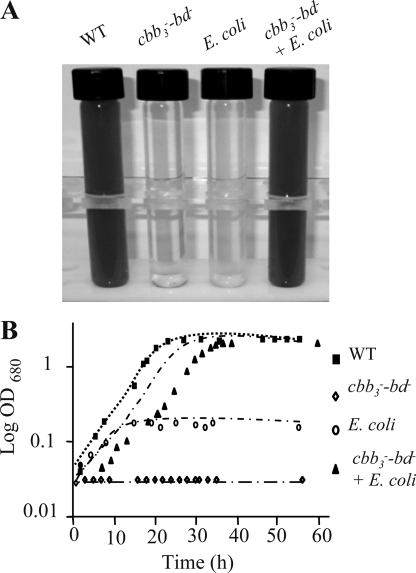
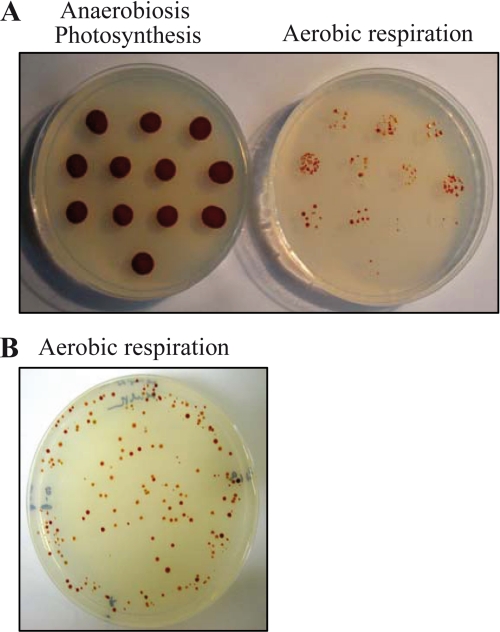
Aerobic growth was affected but still possible in the bd− and cbb3− single deficient mutants. Under aerobic conditions, both strains produced less photosystem but still grew photosynthetically in nondeoxygenated medium. Based on these data, we predicted that aerobic growth and photosystem production would be more affected in the double mutant. Selection of the double mutant cbb3−-bd− was possible only on plates under strict anaerobic photosynthetic conditions in sealed bags with H2- and CO2-producing gas packs. The mutant was respiration-deficient, confirming that cbb3 and bd are the only functional terminal oxidases in R. gelatinosus (supplemental Fig. S2). Interestingly, unlike the wild type and the single mutants, the cbb3−-bd− double mutant cannot carry out photosynthesis in the presence of dissolved O2 in liquid medium (Fig. 3A). Inactivation of both cbb3 and bd gave rise to a strict anaerobe and obligate phototroph.
FIGURE 3.
Cultures (wild type (WT), double mutant cbb3−-bd−, E. coli and the double mutant cbb3−-bd− co-inoculated with E. coli) grown in nondeoxygenated medium under photosynthesis conditions (A) and the corresponding growth curve (B).
Co-inoculation of the cbb3−-bd− mutant with nonphotosynthetic strains like the R. gelatinosus bc1− mutant (data not shown) or E. coli resulted in PS growth of the double mutant as early as 24 h post-inoculation in nondeoxygenated medium. Growth rates of different strains (wild type, cbb3−-bd−, E. coli, and cbb3−-bd− strain co-inoculated with E. coli) grown under photosynthetic conditions were monitored (Fig. 3B). Unlike the double mutant cbb3−-bd−, which did not present any growth, the wild type presented a doubling time of 2.5 h. The E. coli showed a weak growth but stopped at an absorbance of 0.16 in malate medium. The co-inoculated cbb3−-bd− with E. coli presented two phases: the first phase corresponding to E. coli growth, up to 0.16, and the second one after 15 h corresponding to the growth of the double mutant with a growth rate comparable with the wild type.
Survival of the double mutant cbb3−-bd− in liquid medium in the presence of dissolved oxygen under photosynthetic conditions was also monitored. We inoculated several tubes with the cbb3−-bd− strain. Plate count enumeration of cells was carried out each day to check the cell viability. The cell number was stable over 10 days, and no significant cell death was observed. Furthermore, in other cbb3−-bd− tubes, E. coli was added to one culture every 24 h for 10 days (supplemental Fig. S6). The addition of E. coli allowed photosynthetic growth to take place in all tubes, confirming the inhibitory effect of oxygen on photosynthetic growth.
These data show that oxygen had no effect on the long term persistence of the double mutant cells for up to 10 days but inhibits photosynthetic growth. The mutant cells were likely in a “dormant state” in the presence of oxygen and resume photosynthetic growth upon removal of oxygen from the medium by E. coli.
Oxygen Consumption Is a Prerequisite to Initiate Photosynthesis
To monitor the oxygen tension in the culture medium of the wild type, cbb3−-bd− mutant co-inoculated or not with E. coli; cells grown under PS conditions were removed by centrifugation, and the oxygen concentration was measured in the medium. In the anaerobic medium control (nitrogen-sparged malate medium), oxygen concentration was ∼40 nmol/ml. In the nonsparged medium, the oxygen concentration was 250 nmol/ml. In the medium of the cbb3−-bd− mutant, oxygen was not consumed (230 nmol/ml). In contrast, in the growth medium of the PS-grown wild type or cbb3−-bd− co-inoculated with E. coli, the oxygen concentration dropped to 42 nmol/ml. In E. coli growing medium, the oxygen tension also reached 43 nmol/ml. Therefore, as previously suggested, the failure of reducing O2 tension in the medium results in the failure of the initiation of photosynthesis in the double mutant. This is corroborated by the fact that sparging liquid cultures with pure nitrogen gas results in PS growth of the cbb3−-bd− mutant. We thus concluded that medium-dissolved oxygen inhibits PS growth in the cbb3−-bd− mutant.
In R. gelatinosus and likely other anoxygenic phototrophic bacteria, photosynthesis takes place only after the O2 level has reached a minimum. We conclude that in addition to their importance for aerobic growth, the oxidases by depleting the medium from O2 are also required for the start of anaerobic photosynthesis.
caa3 Oxidase as a Suppressor of Respiration and PS Deficiency of the cbb3−-bd− Mutant
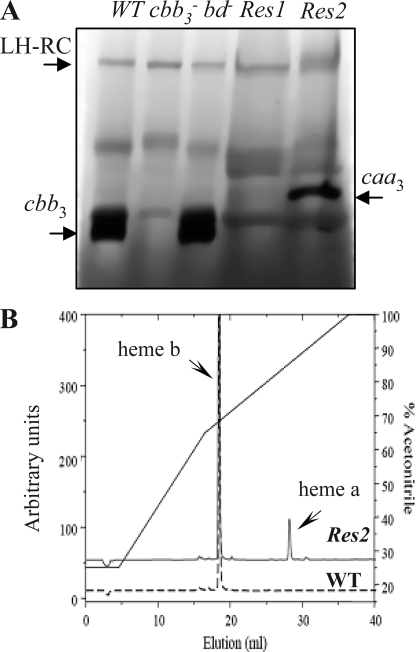
A screen for aerobic growth of the cbb3−-bd− double mutant led to the selection of two suppressor types. In this screen, anaerobic PS-grown cbb3−-bd− cells (108) were spread on plates and incubated under aerobic conditions. Small pigmented colonies appeared 2 weeks later. The newly arising colonies were picked and tested for their respiratory and photosynthetic phenotypes. All of the suppressors regained aerobic growth and simultaneously acquired PS growth ability. The suppressors were also tested for their cytochrome c oxidase activity by the NADI reaction. The majority (50 colonies) of suppressors were NADI− and found to respire with a quinol oxidase. Only two colonies were NADI+, which indicated that the clone contained an active cytochrome c oxidase. Cytochrome c oxidase (TMPD) activities confirmed the presence of cytochrome c oxidase in the suppressor Res2 (50 μmol of TMPDox/min/mg of protein). In the NADI− (suppressor Res1), no TMPD oxidase activity was detected. A search for the mutations in these strains is currently being performed. A detailed account of the suppression mutations in these mutants will be presented elsewhere. We further characterized these strains by in-gel activity assays for the respiratory complexes separated by BN-PAGE. Fig. 4A compares the in-gel cytochrome c oxidase activities in solubilized membranes from different strains of R. gelatinosus grown under semi-aerobic conditions. Both wild type and bd− mutant showed cbb3 oxidase activity (120-kDa band). The oxidase activity was still defective in the Res1 suppressor mutant, whereas the Res2 mutant displayed a slow cytochrome c oxidase activity with an apparent molecular mass of ∼250 kDa. Mass spectroscopy and EPR analyses confirmed that this band corresponds to caa3 cytochrome c oxidase (not shown). Furthermore, HPLC analyses of the wild type and Res2 mutant membranes revealed the presence of heme a only in the Res2 mutant (Fig. 4B). These results support that the Res2 suppressor regained respiration and photosynthesis in nondeoxygenated medium via the caa3 oxidase. As for the cbb3− strain that presented a lag phase following inoculation from a semi-aerobic culture into photosynthesis-permitting conditions, we recorded the photosynthesis growth induction in the Res2 mutant. This strain presented a lag phase of 10 h compared with the wild type (supplemental Fig. S4). This may be explained by the low activity and/or the expression of caa3 oxidase in Res2 resulting in a slow depletion of oxygen from the medium. Taken together these findings confirm that oxygen reductases are essential for photosynthesis start in oxygenated medium in R. gelatinosus and reinforce the dependence of the initiation of photosynthesis on respiration.
FIGURE 4.
Detection of caa3cytochrome c oxidase in Res2 suppressor. A, the active cytochrome c oxidase in BN-PAGE is stained with 3,3′-diaminobenzidine tetrahydrochloride. The cbb3 complex is present in the membrane of the wild type (wt) and of the bd− mutant but not in the cbb3− mutant. The caa3 complex is detected only in the Res2 suppressor. B, noncovalently bound hemes analysis by HPLC from membrane of the wild type and of the Res2 suppressor. Heme b is present in the wild type (WT) and Res2; heme a is present only in the Res2 suppressor.
Insights into the Regulation of Photosystem: PpsR and FnrL Are Critical Factors for Survival in an Aerobic and in an Anaerobic Environment, Respectively
Facultative phototrophs including R. gelatinosus can adapt their metabolism according to oxygen availability and light through aerobiosis or photosynthesis. In R. sphaeroides and R. capsulatus, a complex regulatory network for PS gene regulation in response to changing oxygen concentrations operates according to the intracellular redox state (13, 14). This circuit includes the two-component signal transduction system PrrBA (RegBA), the PpsR and FnrL transcriptional factors. So far, in R. gelatinosus only PpsR and FnrL were identified (16, 20).
PpsR is the main aerobic repressor of photosynthesis genes in purple bacteria, thereby playing a major role in controlling photosystem production under aerobic conditions. If photosynthesis is inhibited in the cbb3−-bd− double mutant in the presence of oxygen because of photosynthesis gene repression by PpsR, then inactivation of ppsR should result in a derepression of photosynthesis even in the absence of the terminal oxidases. To test this hypothesis, we have inactivated ppsR in the double mutant cbb3−-bd− background. The mutant was selected on plates under anaerobic PS conditions and transferred to liquid medium under photosynthetic conditions. The triple mutant was unable to grow by photosynthesis in nondeoxygenated medium, indicating that PpsR was not the only factor that causes inhibition of photosynthesis in the double mutant cbb3−-bd−.
We then asked whether this inhibition was due to an inactivation by oxygen of the FnrL factor. FnrL is a regulator that activates the expression of a variety of genes including photosynthesis genes in response to oxygen limitation in bacteria. Moreover, the FnrL null mutant in R. gelatinosus and R. sphaeroides are nonphotosynthetic (16, 23). In the double mutant cbb3−-bd−, the dissolved O2 might be responsible for the inactivation of FnrL. If this is the case, overexpression of FnrL in the double mutant cbb3−-bd− or the triple mutant cbb3−-bd−-ppsR− may result in derepression of genes encoding proteins involved in photosynthesis function.
Constitutive expression of FnrL in the double mutant did not allow the resumption of photosynthetic growth. However, when FnrL was constitutively expressed in the triple mutant cbb3−-bd−-ppsR−, cells resumed photosynthetic growth in liquid medium. This strain was still unable to grow by respiration, ruling out a possible genetic suppression of the phenotype. With these data we can conclude that PpsR and FnrL are both critical factors for photosynthesis in the presence of oxygen in the medium.
To emphasize the role of oxidases and oxygen regulation on photosynthetic metabolism in R. gelatinosus, a model is presented in Fig. 5. Expression of photosynthesis genes requires the removal of oxygen to abolish gene repression mediated by the PpsR and to induce FnrL-regulated photosynthesis genes. In a high aeration context, reduction of oxygen tension starts with the bd quinol oxidase. When the oxygen tension is sufficiently low, activation of the transcription factor FnrL enhances the expression of cbb3 oxidase (16). Complete reduction of O2 via cbb3 then allows inactivation of PpsR and full activation of the anaerobic regulator FnrL. Under such conditions, expression of photosynthesis genes and photosystem assembly take place to permit anaerobic photosynthetic growth.
FIGURE 5.
Schematic overview of photosynthesis regulation by oxygen in R. gelatinosus. Expression of photosynthesis genes requires the removal of oxygen by reductases and induction of gene expression by FnrL.
On the Role of cbb3 and PpsR in Oxidative Damage: Transcriptional Control of Energy Metabolism through PpsR
It is likely that purple bacteria rely on terminal oxidases and on tight regulation to meet their needs for growth and survival in oxygenated and/or sunny habitat. A close relationship between respiration and oxygen sensors may operate for an optimal regulation of photosynthesis. To address this issue, we inactivate ppsR in the cbb3 or the bd null backgrounds and compare their growth and oxygen tolerance to the ppsR single mutant. The goal is to assess the necessity of these oxidases in the absence of the PpsR repressor. Serendipity has often led to unexpected discoveries. Unlike the bd−-ppsR− mutant that grows aerobically and by photosynthesis in nondeoxygenated medium, the double mutant cbb3−-ppsR− was able to grow by photosynthesis but unable to grow by respiration. Complementation of the mutant in trans either with ppsR or ccoP cloned on a replicative plasmid (PA410 and pBS32, respectively) restores respiration. This suggests that ppsR is somehow involved in bd quinol oxidase expression under aerobic respiration. Interestingly, this mutant was very unstable when grown under photosynthesis in nondeoxygenated liquid medium, but not when grown by photosynthesis under strict anaerobiosis. Cells lacking PpsR are sensitive to photo-oxidative stress caused by an excess of photopigment synthesis in presence of O2. Nevertheless, the ppsR− single mutant is more stable when grown photosynthetically in nondeoxygenated medium thanks to the terminal oxidases. Cells lacking both ppsR and cbb3 are hypersensitive in light and nondeoxygenated medium. This photo-oxidative stress of the cbb3−-ppsR− mutant gave rise to suppressors that recover respiration (Fig. 6) with a very high frequency (10−4). Photosynthetic growth of the cbb3−-ppsR− mutant indicates that bd is active and reduces the oxygen tension to allow photosynthetic growth of the double mutant in nondeoxygenated medium. However, bd quinol oxidase is active in R. gelatinosus under high oxygen tension and is less efficient than cbb3 when oxygen drops. The remaining oxygen in the medium with the excess of photopigment in absence of PpsR should lead to photo-oxidative stress that yields to an unstable strain. In contrast to the bd quinol oxidase, cbb3 is highly induced under microaerobiosis, and its oxygen affinity is very high and should allow complete oxygen depletion in the bd−-ppsR− mutant prior to photosynthesis, explaining its stability when grown photosynthetically in oxygenated medium. These results not only underscore the importance of cbb3 as an oxygen scavenger but also reinforce the relevant role of PpsR as a master regulator of photosystem and its requirement in the protection against photo-oxidative stress when cells are exposed to oxygen and light. This is in agreement with data obtained from R. sphaeroides transcriptome in responses to hydrogen peroxide where down-regulation of the photosynthesis gene is mediated under oxidative stress by the AppA-PpsR regulatory pathway (24).
FIGURE 6.
A, growth of the cbb3−-ppsR− mutant under anaerobic photosynthesis and aerobic respiration. Under aerobic conditions respiratory suppressors begin to appear on plates after a week. B, plate depicting the diversity of suppressors under aerobic conditions.
Conclusion
In this work, we conducted experiments to demonstrate for the first time that terminal oxidases play a crucial role in photosynthesis initiation in the anoxygenic phototrophic bacterium R. gelatinosus. When either cbb3 or bd terminal oxidases were inactivated in R. gelationsus, the resulting mutants were less pigmented and produced lower amounts of photosynthetic complexes. In R. sphaeroides a direct role of the cbb3 in photosynthesis was also reported; but in contrast to R. gelatinosus, the cbb3− mutant from R. sphaeroides was more pigmented than the wild type under aerobic conditions (25). In R. capsulatus, the bc1− mutant like the cbb3− mutant is also more pigmented that the wild type (26, 27). However, the pigmentation is not well pronounced as in R. sphaeroides.
The mechanism whereby disruption of the cbb3 oxidase leads to a derepression of photosynthesis gene expression in presence of O2 in Rhodobacter species remains unclear. In both strains it was suggested that the oxidation state of the quinol pool may be involved in the regulation of photosynthesis gene expression. In R. sphaeroides, it was suggested that the cbb3 oxidase itself is an O2/redox sensor that generates an inhibitory signal transduced through PrrC (Sco1) to the PrrBA (RegBA) two-component system (28). The redox state of the quinone pool is monitored by the redox-active AppA antirepressor protein that controls the function of the PpsR repressor protein. However, in R. capsulatus, PrrC homologue (SenC) was shown to be involved in copper metabolism and in the assembly of a functional cytochrome cbb3 oxidase (29). In this bacterium, although there is no direct evidence that cytochrome cbb3 oxidase directly modulates the activity of the RegB/RegA two-component regulatory system, it was suggested that it is RegB (PrrB) that monitors the redox state of the quinone pool. In the absence of the cbb3 cytochrome oxidase, the excess reduced quinone pool may derepress RegB phosphorylation and lead to the activation of the photosynthesis gene expression under aerobic conditions (30).
When both oxygen reductases (cbb3 and bd) were inactivated in R. gelatinosus, the mutant was unable to grow by photosynthesis even in micromolar concentrations of oxygen. In R. capsulatus, the cbb3−-bd− double mutant has also a peculiar photosynthetic growth defect (it has a growth problem; it would grow much later and revert frequently) in the absence of both oxidases. This mutant also depends on the oxygen availability to establish anoxygenic photosynthetic growth.4
The data presented in this paper suggest that the oxygen level per se is the key element controlling photosynthesis in oxygenated medium in R. gelatinosus and that the role of terminal oxidases is to deplete O2 from the surrounding prior to photosynthesis. Co-inoculation of the mutant with oxygen consuming microorganisms allows the strain to regain photosynthetic growth. This is what probably occurs in nature and in many microbial mats in which aerobes and strict anaerobes coexist and from which axenic in vitro cultivation of strains remains a difficult task. Furthermore, the photosynthetic growth defect of the double mutant cbb3−-bd− provided a tool to select suppressors and confirm the necessary role of oxygen reductases in starting the photosynthetic growth of R. gelatinosus. The caa3 oxidase, although it was not active in the wild type, was required for survival in the suppressor. In this strain, an as yet undefined mutation provides sufficient caa3 oxygen reductase activity to allow the strain to grow by photosynthesis in oxygenated medium. Once again, these results illustrate how photosynthesis and respiration or O2 detoxification are functionally related and emphasize the role of oxygen reductases in photosynthetic growth in this bacterium.
Before the formation of atmospheric oxygen on Earth through oxygenic photosynthesis, life expansion was driven by anaerobic metabolism (31). With oxygen-producing cyanobacteria inducing progressive oxidation of the Earth's atmosphere, the availability of free oxygen would have been growth-inhibiting if not lethal to photosynthetic obligate anaerobes. From an evolutionary perspective, it is tempting to speculate that the ancestral photosynthetic organism may have been a strict anaerobe in agreement with the work of Tice and Lowe (32) that evolved later to a phototroph and aerobe upon the release of oxygen in the Earth's atmosphere and the acquisition of oxygen reductases. The oxygen reductases or O2 scavengers probably evolved to provide first oxygen detoxification and then to support evolution of the aerobic world.
Supplementary Material
Acknowledgments
We acknowledge discussions with M. Picaud, A. Durand, L. Sperling, and M. Usdin. Thanks to W. Nitschke for EPR data and helpful discussion.
This work was financially supported by French Agence Nationale de la Recherche Grant ANR BLAN06-2_147814).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
B. Khalfaoui Hassani, R. van Lis, A.-L. Ducluzeau, I. Agalidis, U. Liebl, C. Astier, W. Nitschke, S. Ouchane, and B. Schoepp-Cothenet, manuscript in preparation.
F. Daldal, personal communication.
- PS
- photosynthesis
- Km
- kanamycin
- BN
- blue native
- HPLC
- high pressure liquid chromatography
- NADI
- N′,N′-dimethyl-p-phenylenediamine
- TMPD
- tetramethyl-p-phenylenediamine
- RT
- reverse transcription
- qPCR
- quantitative real time RT-PCR.
REFERENCES
- 1.Allen J. F., Martin W. (2007) Nature 445, 610–612 [DOI] [PubMed] [Google Scholar]
- 2.Falkowski P. G. (2006) Science 311, 1724–1725 [DOI] [PubMed] [Google Scholar]
- 3.Raymond J., Blankenship R. (2004) Geobiology 2, 199–203 [Google Scholar]
- 4.Raymond J., Segrè D. (2006) Science 311, 1764–1767 [DOI] [PubMed] [Google Scholar]
- 5.Tomitani A., Knoll A. H., Cavanaugh C. M., Ohno T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5442–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones K. M., Haselkorn R. (2002) J. Bacteriol. 184, 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valladares A., Herrero A., Pils D., Schmetterer G., Flores E. (2003) Mol. Microbiol. 47, 1239–1249 [DOI] [PubMed] [Google Scholar]
- 8.Steunou A. S., Jensen S. I., Brecht E., Becraft E. D., Bateson M. M., Kilian O., Bhaya D., Ward D. M., Peters J. W., Grossman A. R., Kühl M. (2008) Isme J. 2, 364–378 [DOI] [PubMed] [Google Scholar]
- 9.Zufferey R., Preisig O., Hennecke H., Thöny-Meyer L. (1996) J. Biol. Chem. 271, 9114–9119 [DOI] [PubMed] [Google Scholar]
- 10.Poole R. K., Hill S. (1997) Biosci. Rep. 17, 303–317 [DOI] [PubMed] [Google Scholar]
- 11.Baughn A. D., Malamy M. H. (2004) Nature 427, 441–444 [DOI] [PubMed] [Google Scholar]
- 12.Frank H. A., Cogdell R. J. (1996) Photochem. Photobiol. 63, 257–264 [DOI] [PubMed] [Google Scholar]
- 13.Zeilstra-Ryalls J. H., Gomelsky M., Yeliseev A. A., Eraso J. M., Kaplan S. (1998) Methods Enzymol. 297, 151–166 [DOI] [PubMed] [Google Scholar]
- 14.Bauer C., Elsen S., Swem L. R., Swem D. L., Masuda S. (2003) Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouchane S., Steunou A. S., Picaud M., Astier C. (2004) J. Biol. Chem. 279, 6385–6394 [DOI] [PubMed] [Google Scholar]
- 16.Ouchane S., Picaud M., Therizols P., Reiss-Husson F., Astier C. (2007) J. Biol. Chem. 282, 7690–7699 [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 18.Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 19.Lemaire C., Dujardin G. (2008) Methods Mol. Biol. 432, 65–81 [DOI] [PubMed] [Google Scholar]
- 20.Steunou A. S., Astier C., Ouchane S. (2004) J. Bacteriol. 186, 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whelan J. A., Russell N. B., Whelan M. A. (2003) J. Immunol. Methods 278, 261–269 [DOI] [PubMed] [Google Scholar]
- 22.Ouchane S., Nitschke W., Bianco P., Vermeglio A., Astier C. (2005) Mol. Microbiol. 57, 261–275 [DOI] [PubMed] [Google Scholar]
- 23.Zeilstra-Ryalls J. H., Gabbert K., Mouncey N. J., Kaplan S., Kranz R. G. (1997) J. Bacteriol. 179, 7264–7273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeller T., Moskvin O. V., Li K., Klug G., Gomelsky M. (2005) J. Bacteriol. 187, 7232–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeilstra-Ryalls J. H., Kaplan S. (1996) J. Bacteriol. 178, 985–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daldal F., Davidson E., Cheng S. (1987) J. Mol. Biol. 195, 1–12 [DOI] [PubMed] [Google Scholar]
- 27.Koch H. G., Hwang O., Daldal F. (1998) J. Bacteriol. 180, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh J. I., Kaplan S. (2001) Mol. Microbiol. 39, 1116–1123 [DOI] [PubMed] [Google Scholar]
- 29.Swem D. L., Swem L. R., Setterdahl A., Bauer C. E. (2005) J. Bacteriol. 187, 8081–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swem L. R., Gong X., Yu C. A., Bauer C. E. (2006) J. Biol. Chem. 281, 6768–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canfield D. E., Rosing M. T., Bjerrum C. (2006) Philos. Trans. R. Soc. Lond B. Biol. Sci. 361, 1819–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tice M. M., Lowe D. R. (2004) Nature 431, 549–552 [DOI] [PubMed] [Google Scholar]
- 33.Uffen R. L. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 3298–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prentki P., Krisch H. M. (1984) Gene 29, 303–313 [DOI] [PubMed] [Google Scholar]
- 35.Kovach M. E., Phillips R. W., Elzer P. H., Roop R. M., 2nd, Peterson K. M. (1994) BioTechniques 16, 800–802 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.