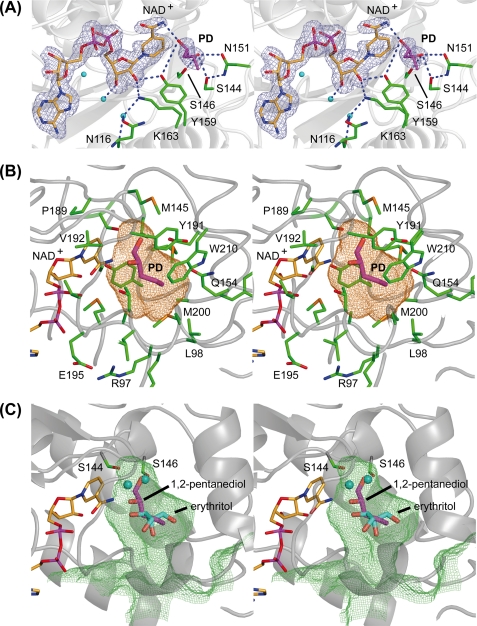
FIGURE 4.
Stereo representation of the substrate binding pocket of GatDH. The main-chain trace of the protein is displayed as a ribbon in gray. The carbon atoms of the cofactor NAD+ are colored in orange, and those of the amino acid residues are in green. One-letter codes are used to identify amino acids. A, electron density map around the cofactor and the substrate 1,2-(S)-pentanediol (PD, carbon atoms in magenta). The final σA-weighted (2Fobs − Fcalc) electron density omit map drawn in blue and contoured at 2σ was calculated after removal of the substrate from the model. Side chains in the surrounding neighborhood are displayed as sticks. The blue dashed lines mark the coordinating hydrogen bonds related to substrate binding and the catalyzed redox reaction. B, stick representation of the cofactor NAD+ and substrate 1,2-(S)-pentanediol (PD) superimposed with the mesh representation of the substrate binding pocket. The substrate binding pocket was analyzed with the program VOIDOO (62) and displayed with PyMOL (36). The cavity is displayed in a mesh representation and colored in orange for a probe radius of 1.4 Å (inner part of the binding pocket). The side chain of those amino acids that define the substrate binding pocket are displayed as sticks and are superimposed on the cofactor, the substrate, and the binding cavity. C, superposition of meso-erythritol (carbon atoms in cyan) together with its two coordinating water molecules (cyan spheres) onto the structure of GatDH with bound 1,2-(S)-pentanediol (carbon atoms in magenta) and the mesh representation of the substrate binding pocket in lime green for a probe radius of 1.1 Å (indicating the access path of the substrate). The two serine residues Ser144 and Ser146 are represented as sticks indicating their importance for the positioning of the substrate (in the case of 1,2-(S)-pentanediol) or the well coordinated water molecules within the substrate binding pocket. For clarity, the other amino acid residues of the binding pocket are not shown.