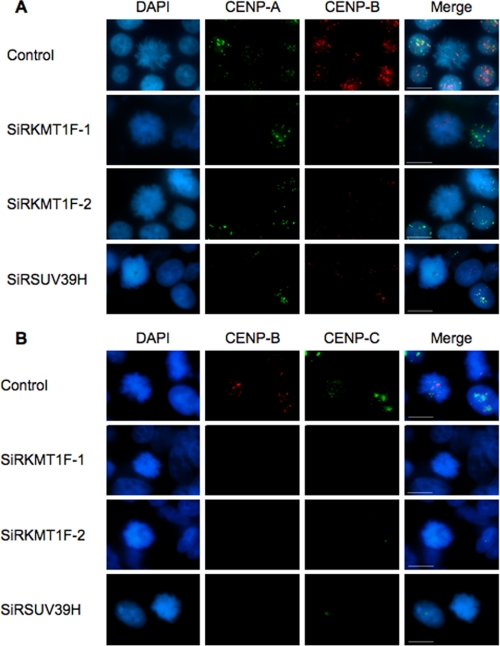
FIGURE 3.
KMT1F depletion correlates with decreased CENP B and C staining at mitotic chromosomes. To further investigate the link between KMT1F and CREST staining, we depleted KMT1F or SUV39H1 in 293T cells and analyzed CENP A, CENP B, or CENP C staining by indirect immunofluorescence in mitotic cells using either mouse anti-CENP A (1/250; Abcam), rabbit anti-CENP B (1/100; Abcam), or mouse anti-CENP C (1/250; Abcam) for 2 h. Secondary anti-mouse and anti-rabbit antibodies were used for detection as previously described. The cells were counterstained with DAPI (Sigma). Imaging of 30–50 mitotic cells was performed using conventional microscopy. A, costaining of CENP A and B. B, costaining of CENP B and C. Scale bars, 10 μm. In KMT1F-depleted cells, CENP A staining is not significantly modified, whereas CENP B (p < 0.001 for SiRKMT1F-1 (58% of cells with loss of CENP B signal) or SiRKMT1F-2 (79% of cells with loss of CENP B signal versus control using a Student's t test)) and CENP C staining are significantly decreased in mitotic cells (p < 0.001 for SiRKMTF-1 (63% of cells with loss of CENP C signal) or SiRKMTF-2 (90% of cells with loss of CENP B signal) versus control using a Student's t test)). Similar results were obtained after depletion in SUV39H (p < 0.001, 86% of cells with loss of CENP B signal or 76% of cells with loss of CENP C signal versus control using a Student's t test), suggesting that both histone methyltransferases contribute to the trimethylation of H3K9me3 residues at centromeres and thereby regulate the assembly of functional centromeres.