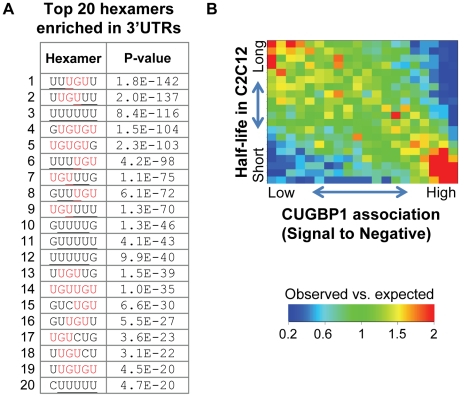
Figure 5. CUGBP1 bound mRNAs contain GREs in their 3′UTRs.
(A) Top 20 ranked significant hexamers in the 3′UTRs of mRNAs with signal-to-negative ratios above 95th-percentile. P-values were derived from Fisher's exact test comparing frequency of occurrence in top 5% transcripts with that in other transcripts. Consecutive Us > = 3 are underlined, and UGU is shown in red. (B) Comparison of signal-to-negative ratios in the CUGBP1 RIP-Chip experiment with mRNA half lives in C2C12 cells. A gene density plot was used to show the relationship, in which mRNAs were evenly divided into 20 groups based on signal-to-negative ratio (x-axis) or half life (y-axis). The number of mRNAs in each cell of the 20×20 table (observed value, obs) was normalized to the mean of all cells (expected value). The ratios (observed/expected) are shown in a heat map according to the color scale shown in the figure. Red represents enrichment, and Blue for depletion.