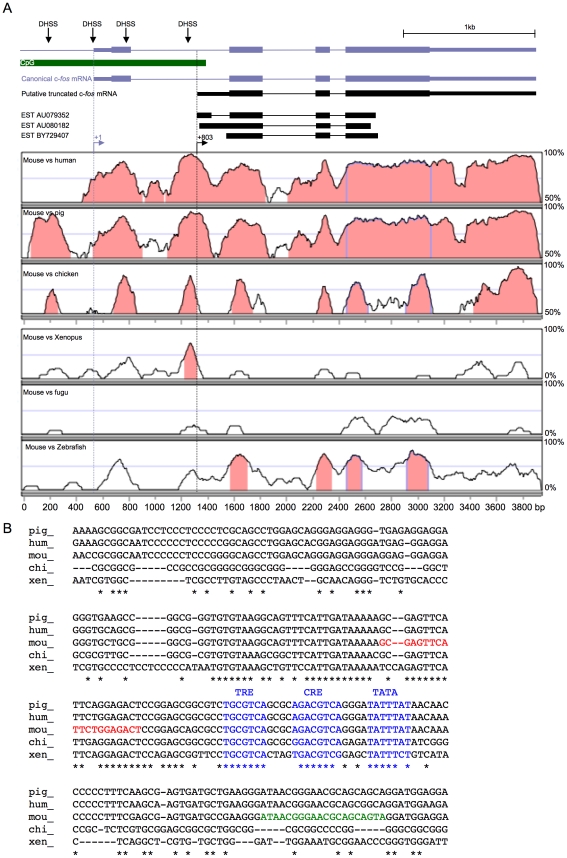
Figure 1. An extremely conserved region in c-fos first intron: evidence for a nested promoter.
A. c-fos gene conservation profile between the mouse gene and 6 other indicated species (VISTA genome server). The baseline corresponds to 50% identity for human, pig and chicken genomes, and to 0% identity for xenopus, fugu, and zebrafish genomes in 100 bp windows. Peaks culminating at more than 70% identity (in 100 bp windows) are painted in pink by the software. Note that the 3′ half of first intron is more conserved than surrounding exons in the four top species. Confirmed start site for canonical mRNA and putative intronic mRNA appear respectively as blue and black broken arrows. A CpG island (green box) extends from the canonical promoter to the putative intronic promoter. Blue boxes represent exons, black mRNAs correspond to the putative mRNA starting in the conserved region and three ESTs previously sequenced and mapping to the same region (UCSC genome server). DNase I hypersensitive sites from Renz et al. are shown as black downward arrows. B. Nucleotide alignment of the most conserved part of c-fos first intron (from +619 to +849 relative to mouse canonical TSS) from 5 species (pig, human, mouse, chicken, xenopus). Asterisks depict nucleotides identical in all 5 species. Conservation culminates in motifs resembling a TRE, a CRE, and a TATA box (shown in blue). Red and green sequences respectively correspond to primers 1 and 2 used in figure 6.