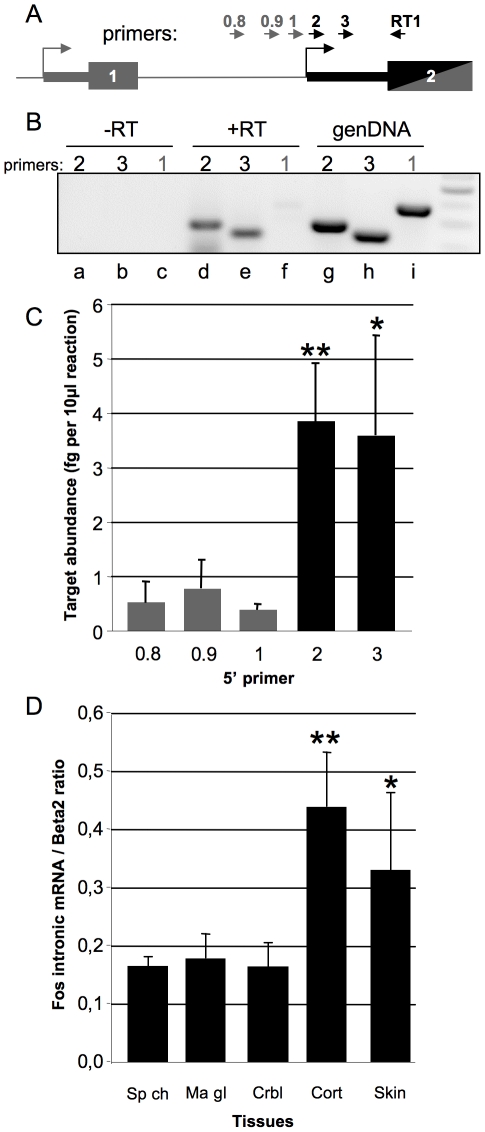
Figure 6. Validation of the expression profile and start site region of c-fos intronic mRNA by RT-QPCR.
A. Schematic representation of the primers used in this RT-PCR study. Boxes represent exons, larger boxes are translated regions, arrows depict primers. The 3′ PCR primer was always RT1. 5′ primers in grey lie upstream the putative TSS, whereas primers in black are downstream. B. RT-PCR mapping of the c-fos intronic mRNA start site region. RNA extracted from E12.5 mouse embryo mammary buds was reverse transcribed with primer RT1, then cDNAs were amplified with primers used for detection of the intronic mRNA (primer 2 lane d, primer 3 lane e) and canonical pre-mRNA (primer 1, lane f). Minus RT control ensures that no genomic DNA contaminant was amplified with any primer (-RT: lanes a, b, c), while PCR on genomic DNA (genDNA: lanes g, h, i) shows that primer 1 is PCR-competent (lane i). The DNA ladder (last lane) shows fragments of 600, 500, 400, 300, and 200 bp. n = 4. C. Quantitative PCR-mediated mapping of the intronic mRNA start site on cDNA from adult mouse cortex. The five different 5′ primers described earlier were used along with RT1 for amplification. Signal was normalized according to standard curves elaborated for each primer pair, allowing a measure of the targeted RNA independently of primer efficiency. The scale shows the amount of RNA in fg per 10 µl of reaction. Error bars represent standard deviation, n = 4. D. RT-QPCR analysis of fos intronic mRNA expression in adult tissues. Quantitative PCR was ran with primers 2 and RT1 on cDNA prepared from dissected candidate adult mouse tissues (from left to right: spinal chord, mammary gland, cerebellum, cortex, skin). Normalization was done over the expression of beta-2 microgobulin. Error bars represent standard deviation, n = 4. Statistical analysis of variance (ANOVA) was performed relative to primer 0.8 (C) or to spinal chord level (D). *: p<0.05; **: p<0.01.