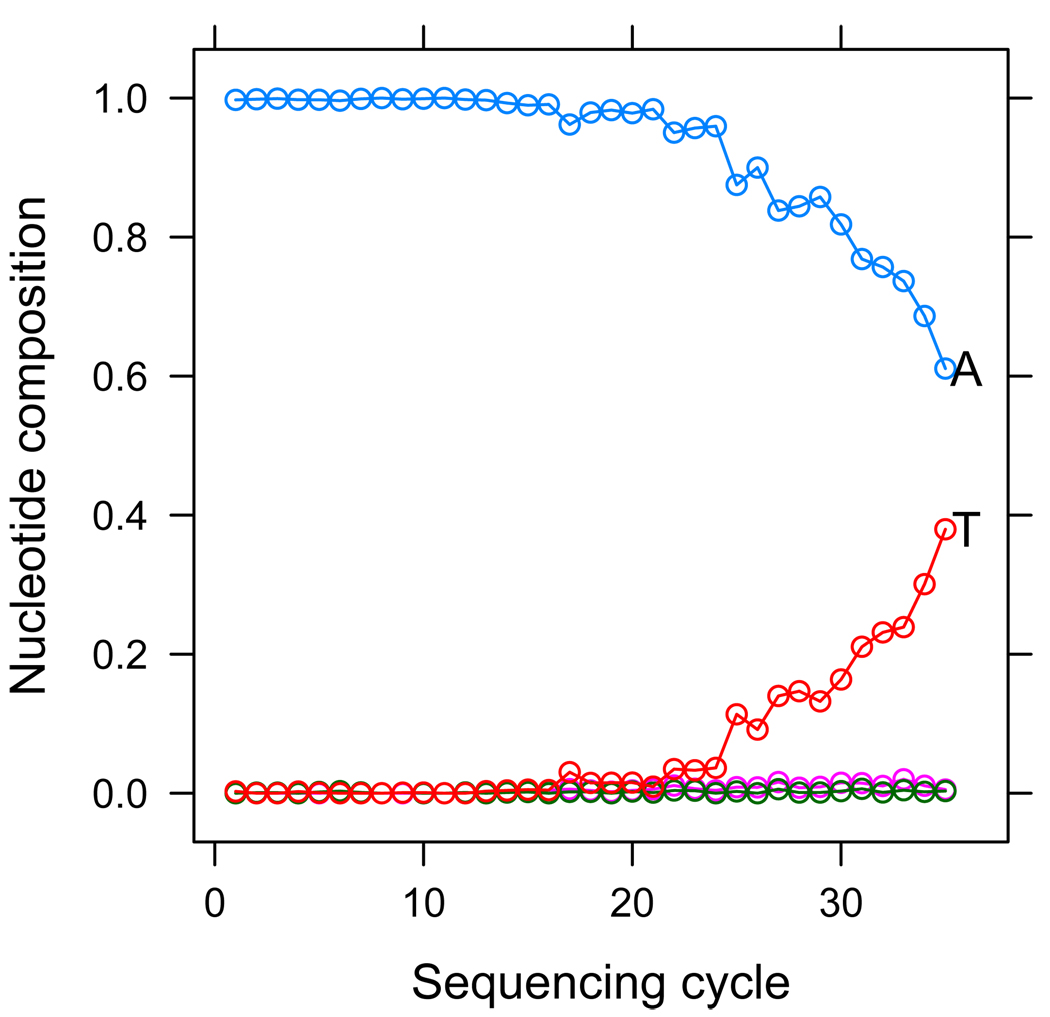
Figure 2.
We plot the base composition of reads aligning to a specific genomic position falsely reported by MAQ as a SNP. We stratify base composition by cycle, that is, at tick 1 of the x-axis we plot the base composition of the first cycle in reads which align to the SNP site in the first cycle, and so on. We can see a trend whereupon T’s become much more frequent only in reads that align to the SNP site at later sequencing cycles, indicating a technical bias in base-calls at this position.