Fig. 1.
(A) IS trajectory. Two sample windows of the IS energies of the GS10 peptide in kcal/mol and the room temperature potential energy (in arbitrary units for comparison) time series are shown in red and gray, respectively. In the left part a transition between the 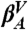 and
and 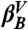 valley is depicted, whereas in the right part a series of transitions between multiple valleys is shown. (B) CSN of the GS10 peptide. Nodes and links represent IS and MD transitions, respectively; the color code corresponds to that in D, except that white nodes are characterized by Z > 0.97. The size of the nodes and links is proportional to their populations. For clarity, only nodes that have been visited more than 200 times are shown; there are a total of 571 nodes. (C) IS CFEP of the GS10 peptide relative to the most populated microstate, βA. The four most populated valleys are shown in dashed lines, and the corresponding lowest energy microstate structures are schematically sketched; only the atoms involved in the relevant hydrogen bonding and SER orientations are displayed. (D) Reaction pathways between the most important IS of the system displayed as a CCSN. IS populations and average number of transitions are shown both by the numbers and by the size of the nodes and the thickness of the links, respectively (see text for comments).
valley is depicted, whereas in the right part a series of transitions between multiple valleys is shown. (B) CSN of the GS10 peptide. Nodes and links represent IS and MD transitions, respectively; the color code corresponds to that in D, except that white nodes are characterized by Z > 0.97. The size of the nodes and links is proportional to their populations. For clarity, only nodes that have been visited more than 200 times are shown; there are a total of 571 nodes. (C) IS CFEP of the GS10 peptide relative to the most populated microstate, βA. The four most populated valleys are shown in dashed lines, and the corresponding lowest energy microstate structures are schematically sketched; only the atoms involved in the relevant hydrogen bonding and SER orientations are displayed. (D) Reaction pathways between the most important IS of the system displayed as a CCSN. IS populations and average number of transitions are shown both by the numbers and by the size of the nodes and the thickness of the links, respectively (see text for comments).

