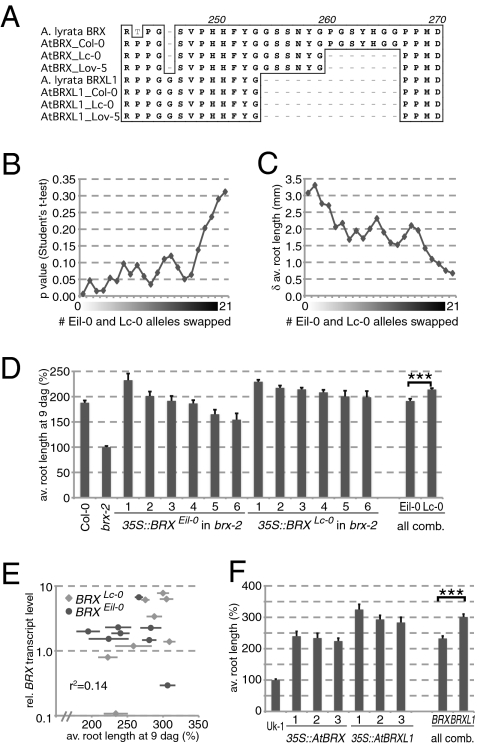
Fig. 3.
Quantitative functional analyses of the AtBRXLc-0 allele. (A) Alignment of the homologous spacer 3 regions of AlBRX, AlBRXL1, AtBRX, and AtBRXL1, the latter including the Col-0, Eil-0, and Lc-0 alleles. (B) Analysis of recombinant inbred lines derived from an Eil-0 × Lc-0 cross. Significance of average root length difference between 49 lines that carry the Lc-0 allele of AtBRX vs. 57 lines that carry the Eil-0 allele (at zero on x axis), and increasing permutation by swapping lines between the two groups. (C) As in B, for absolute difference in root length. (D) Average propensity of the AtBRXEil-0 and AtBRXLc-0 alleles to complement the Arabidopsis brx-2 null mutant short root phenotype when expressed under control of the 35S promoter in transgenic brx seedlings, scored at 9 dag. The six best rescuing independent transgenic lines for each construct are shown, as well as all data combined. (E) Correlation graph of BRX expression level (log) vs. root length (percentage), determined in a replicate experiment similar to D, with all eight independent transgenic lines. BRX mRNA levels were determined by quantitative PCR, expressed as relative expression level as compared with the housekeeping gene, EF1 (15). For reference, expression level in Col-0 wild-type control was 0.0021 ± 0.0010. (F) As in D, for AtBRX and AtBRXL1 expressed in the brx null mutant background, Uk-1, three independent transgenic lines each.