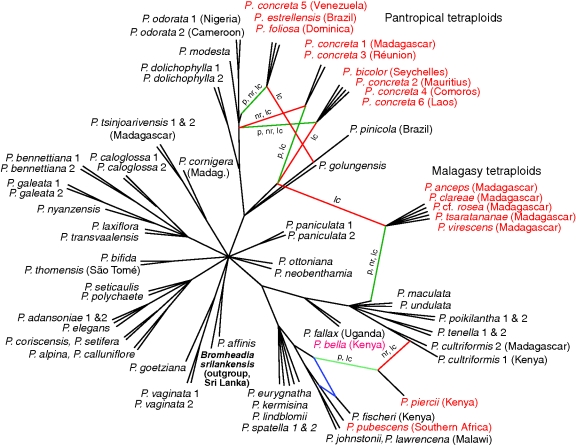
Fig. 6.
Unrooted consensus galled network (40 % threshold for network construction) summarizing incongruities and allopolyploids in the five individual gene trees of diploid and tetraploid accessions, using 50 % bootstrap consensus trees as input. Branch lengths are not to scale; only the topology is shown. Reticulations inferred solely from incongruence between the gene trees are coloured blue. Red- and green-coloured branches represent reticulations involving cloned tetraploids, for which the parental sequences were manually reconnected as hybridization nodes. Green branches represent relationships according to the plastid DNA tree; red branches are only present in nuclear DNA trees; these branches are also annotated if they are represented in the plastid (p), ITS (nr) or nuclear low-copy (lc) gene trees. Species names of the cloned tetraploid samples are coloured red. Accessions are from mainland tropical Africa unless otherwise indicated.