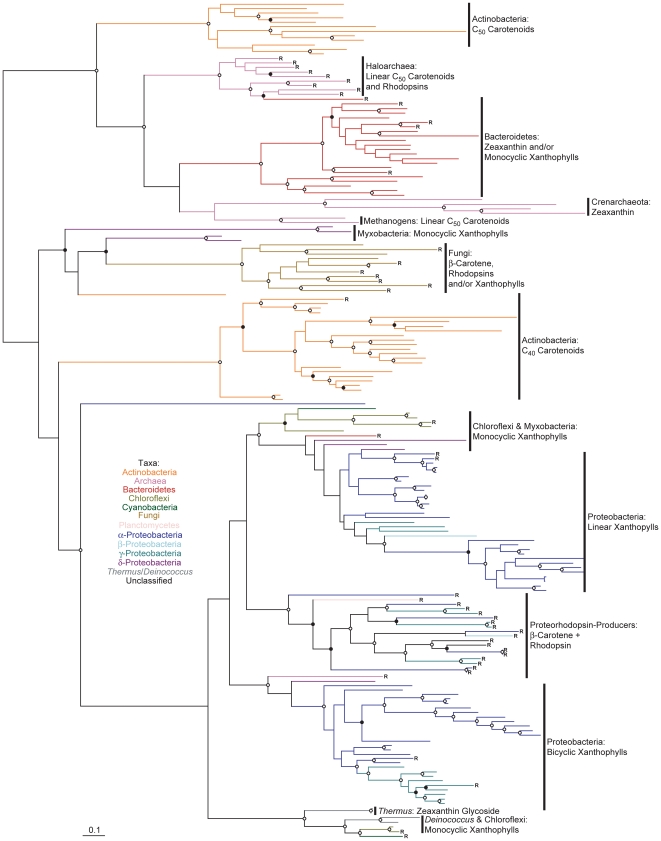
Figure 3. Phylogenetic tree of CrtI protein sequences constructed using RAxML.
Bootstrap values are indicated as a percentage of the automatically determined number of replicates determined using the CIPRES web portal; those ≥80% are indicated by an open circle and those ≥60% but <80% by a filled circle. For a version of this tree containing sequence names and numerical bootstrap values see Figure S2. Genomes containing a rhodopsin homolog are indicated by an “R”. Carotenoids typical of each lineage are indicated to the right of each clade; note that not all structures are included. The scale bar represents 10% sequence divergence. The tree is rooted to its midpoint to maximize the clarity of intraclade relationships.