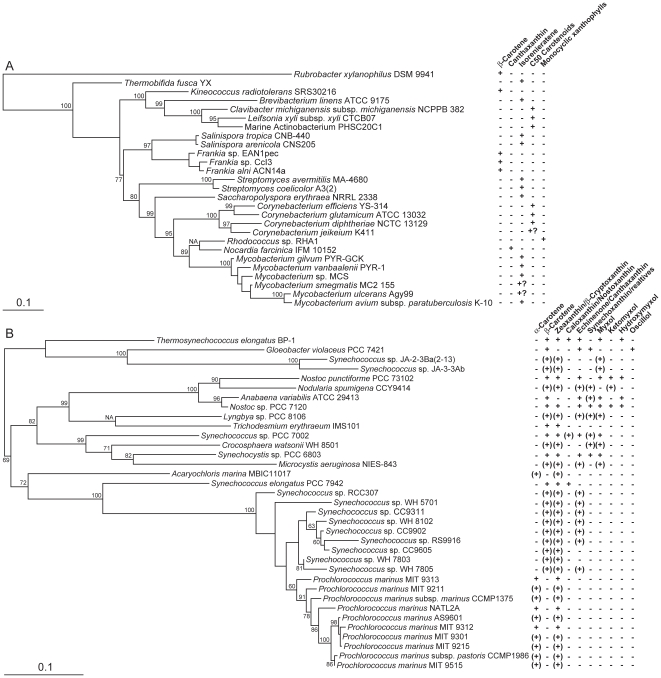
Figure 8. Phylogenetic trees constructed from nearly full-length 16S rRNA genes from carotenoid-producing members of (A) Actinobacteria and (B) Cyanobacteria constructed using RAxML.
Bootstrap values ≥60% are indicated as a percentage of the automatically determined number of replicates determined using the CIPRES web portal. All trees are rooted to their midpoint, and the scale bar represents 10% sequence divergence. NA indicates the ML basal node for which no bootstrap value was given. Question marks indicate organisms for which carotenoid biosynthetic pathways are incomplete, likely from genomic decay. For Cyanobacteria, known carotenoids are derived from the compilations of Maresca et al. [31] and Takaichi and Mochimaru [33], with inferences derived from in silico pathway reconstructions (Table S1) indicated in brackets. For Actinobacteria, carotenoid pathway products are nearly exclusively derived from pathway reconstructions (Table S1) due to the lack of 16S rRNA genes for most biochemically studied strains. Note that for clarity, not all terminal pathway modifications (especially glycosylations) are indicated, and carotenoids similarly modified at each end are grouped together because of the difficulty in determining this level of substrate specificity via exclusively in silico analysis.