Abstract
The recently discovered pneumococcal serotype 6C was created when the original wciN gene in the 6A capsule gene locus was naturally replaced with a new gene. Since the capsule gene loci of 6A and 6B serotypes may differ by only one base pair in the wciP gene, it was speculated that a new serotype ‘6D’ would be possible if the new wciN gene were inserted into the 6B capsule gene locus. Although pneumococci expressing serotype 6D could be produced in the laboratory, initial searches for natural pneumococcal isolates expressing serotype 6D were unsuccessful. However, we now report the discovery of two naturally occurring pneumococcal isolates from Korea that have the serological, genetic and biochemical features predicted for serotype 6D.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is a significant human pathogen (Fedson, 1988), and expresses on its surface serotype-specific capsular polysaccharides (PSs), which greatly increase its virulence (Avery & Dubos, 1931). However, antibodies to the capsule can abrogate the virulence and provide serotype-specific protection (Cole, 1913). S. pneumoniae as a species can produce more than 90 different capsule types (Park et al., 2007b), but not all capsule types are equally prevalent in human diseases. For instance, prior to the use of conjugate vaccines, several serotypes such as 6A, 6B, 14, 19A, 19F and 23F were responsible for a large number of invasive pneumococcal diseases (Robinson et al., 2001) and consequently have been extensively studied.
Serotypes 6A and 6B have been grouped into serogroup 6 because of their serological similarities. Also, serotypes 6A and 6B consistently differ in their 17 kb capsule gene loci by only one nucleotide in the wciP gene (Mavroidi et al., 2004). This difference produces 6A WciP with a serine at residue 195 but 6B WciP with an asparagine at the same residue (Mavroidi et al., 2004). Serotypes 6A and 6B produce capsular PSs with almost identical structures except for a different rhamnose–ribitol linkage (Kamerling, 2000). Recently, we discovered a new member of serogroup 6 which we named 6C (Park et al., 2007b). Serotype 6C is distinguished serologically from serotypes 6A and 6B by its unique monoclonal antibody (mAb) binding profile (Lin et al., 2006). Subsequent genetic studies have shown that the capsule gene loci of serotypes 6A and 6B have wciN6A (named alternatively as wciNα here), but that the 6C capsule gene locus has wciN6C (or wciNβ) (Park et al., 2007a). Finally, biochemical studies revealed that 6C PS has a glucose residue in place of the galactose residue present in the 6A or 6B PSs (Park et al., 2007b).
Following the discovery of serotype 6C, we postulated the presence of a new serotype, which could be created by mutating the critical nucleotide in the wciP gene of the 6C capsule gene locus or by inserting the wciNβ gene into the 6B capsule gene locus (Bratcher et al., 2009). This new serotype, which logically could have been named 6D, was provisionally named 6X1 because the new serotype had not been found in nature (Bratcher et al., 2009). Nevertheless, our group continued the search and discovered natural isolates expressing serotype 6D in Korea; we describe our findings below.
METHODS
Bacterial strains and culture.
TIGR6A, TIGR6B, TIGR6C and TIGR6X1, which are isogenic strains of TIGR4 expressing the four different capsule serotypes of serogroup 6, were used as controls for the assays (Bratcher et al., 2009). Fourteen serogroup 6 isolates, including MNZ21 and MNZ22, were obtained from the nasopharyngeal cultures of 14 healthy children (less than 5 years old) attending a day-care centre in Jeju Island in Korea in 2008 and were subsequently transferred to the Nahm laboratory for further analysis. These isolates showed the typical α-haemolytic colony morphology of S. pneumococcus on blood agar plates, were susceptible to optochin, and were bile soluble. All bacteria were grown in Todd–Hewitt broth (BD Biosciences) supplemented with 0.5 % yeast extract (THY) and kept frozen at −80 °C until used.
Flow cytometry.
Aliquots of frozen bacteria were thawed, washed, resuspended in FACS buffer (PBS containing 3 % FBS and 0.1 % sodium azide) and incubated with culture supernatants of hybridomas (diluted 1 : 10 in FACS buffer) for 20 min at room temperature with shaking. After washing, the bacteria were incubated with fluorescein-conjugated goat antibody to mouse immunoglobulin for 20 min at room temperature with shaking. After washing away unbound goat antibody, the bacteria were resuspended in FACS buffer containing Syto9 (160 nM) and examined with a flow cytometer (FACSCalibur, Becton Dickinson). The data were then analysed with the Cell Quest program. Isotype-matched negative controls were used to identify negative staining and their fluorescence signals were less than 20 units (data not shown).
PCR and DNA sequencing.
PCR mixtures contained 38.8 μl sterile water, 2 μl of each 5 pmol μl−1 primer, 2 μl 10 mM dNTPs, 5 μl 10× buffer solution and 0.2 μl LA Taq polymerase (2.5 U μl−1, Takara Biomedical). For the template, either chromosomal DNA isolated with the Wizard genomic DNA purification kit (Promega) or colonies grown on blood agar plates were used. Thermal cycling conditions were previously described (Park et al., 2007a). PCR products were analysed by electrophoresis in 1 % agarose gel. The primers used here were the forward primer for wciN (5106), TACCATGCAGGGTGGAATGT; the reverse primer for wciN (3101), CCATCCTTCGAGTATTGC; the forward primer for wciP (5108), ATGGTGAGAGATATTTGTCAC; and the reverse primer for wciP (3107), AGCATGATGGTATATAAGCC. PCR products were purified using the Wizard PCR Clean-up System (Promega), and the DNA sequencing was performed by the genomics core facility at the University of Alabama at Birmingham. DNA sequences were analysed with Lasergene v. 5.1 software (DNASTAR).
Inhibition ELISA.
Capsular PSs were distinguished using an inhibition-type ELISA. Briefly, the wells of ELISA plates (Corning Costar Corp.) were coated at 37 °C with 5 μg ml−1 of 6B capsular PS (ATCC, Manassas, VA, USA) overnight in PBS. After washing the plates three times with PBS containing 0.05 % Tween 20, 50 μl of a previously diluted bacterial culture supernatant (or lysate) was added to the wells along with 50 μl of an anti-6B mAb. Pneumococcal lysates were prepared by growing pneumococci overnight in 1 ml THY broth without shaking and then incubating the tubes for 15 min at 37 °C with 110 μl of a lysis buffer (0.1 % sodium deoxycholate, 0.01 % SDS and 0.15 M sodium citrate in deionized water). Culture supernatants of 6B-specific hybridomas Hyp6BM7 and Hyp6BM8 were used at dilutions of 1 : 50 and 1 : 100, respectively. These hybridomas were produced from the fusion of myeloma cells with spleen cells isolated from mice immunized with 6B PS (Sun et al., 2001). After 30 min incubation in a humid incubator at 37 °C, the plates were washed three times and incubated for 30 min with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin (Sigma). The plates were washed three times. Then 100 μl p-nitrophenyl phosphate substrate (Sigma) in diethanolamine buffer at a concentration of 1 mg ml−1 was added, and the plates were allowed to incubate at room temperature for 1–2 h so that an optimal absorbance (maximal A405 of 1.5) was reached. The A405 was read with a microplate reader (BioTek Instruments).
Hydrolytic stability assay.
A 0.9 ml sample of 2 mg ml−1 PS in water was mixed with 0.1 ml 0.1 M NaOH. This solution was then divided between two Eppendorf tubes and incubated at 85 °C. Capsular PSs from serotypes 6A, 6B, 6C and 6X1 were described before (Bratcher et al., 2009). Capsular PS from a 2 l culture of MNZ21 was purified using ethanol precipitation after lysis and removal of cell debris as previously described (Park et al., 2007b). The precipitate was then subjected to molecular mass sizing chromatography (Sephacryl S-500HR, 500 ml column volume) and fractions positive for PS were pooled and lyophilized. At the indicated times, 0.1 ml was removed from these samples, neutralized with 0.1 M HCl and then stored at 4 °C until use in the inhibition ELISA. Using the same buffers and incubation conditions as described for the inhibition ELISA above, plates were coated with 100 μl of 5 μg ml−1 6A, 6B, 6C, 6X1 or MNZ21 PS (Bratcher et al., 2009). The ELISA was performed with the hydrolysed samples on plates coated with their respective PSs. For 6A and 6C PSs, Hyp6AG1 was used as the primary antibody (as performed by Park et al., 2007b); for 6B and MNZ21 PSs, Hyp6BM8 was used (as described above). Data shown are the mean of samples run in duplicate.
Monosaccharide composition analysis of PS.
Capsular PS from MNZ21 was purified as described above. 6B PS from ATCC was used as a control. A 15 μg sample of lyophilized capsular PS was dissolved in 400 μl 1.5 M methanolic HCl and incubated at 80 °C for 16 h. After evaporating the methanolic HCl in a vacuum centrifuge, the sample was redissolved in 250 μl methanol, transferred to a glass GC vial insert and again dried in a vacuum centrifuge. The sample was then trimethylsilylated with 50 μl Tri-Sil Reagent (Pierce Biotech). The reaction products were analysed as described by Baker et al. (2006) using a gas–liquid chromatograph (HP5890, Hewlett Packard) fitted with a 30 m HP-1 wide-bore fused-silica column coated with a 0.88 μm layer of cross-linked methylsilicone gum. The column temperature was maintained at 100 °C for 5 min and then increased to 275 °C at a rate of 20 °C min−1. Finally, it was held at 275 °C for 5 min.
RESULTS AND DISCUSSION
Serological findings
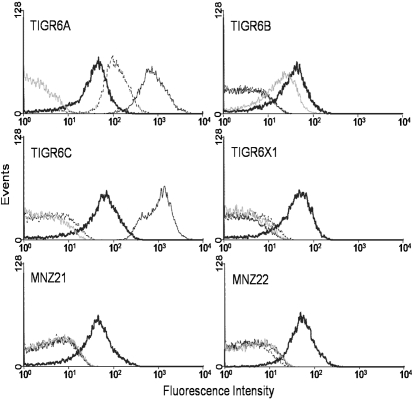
Studies with TIGR6X1 showed that it binds to a mAb (Hyp6BM8), but that it does not react with other mAbs reacting with serotypes 6A, 6B and 6C (Bratcher et al., 2009). We used flow cytometry to test the two isolates (MNZ21 and MNZ22) and four laboratory strains expressing serotypes 6A, 6B, 6C and 6X1 for their ability to bind to this panel of mAbs. As shown in Fig. 1, we were able to directly demonstrate that MNZ21, MNZ22 and TIGR6X1 bound to Hyp6BM8, but did not bind to the three other mAbs. In contrast, the three control strains bound Hyp6BM8 (thick black line) as well as at least one of the three other mAbs [Hyp6AM3 (dashed line) stained TIGR6A, Hyp6AG1 (thin black line) stained TIGR6A and TIGR6C, and Hyp6BM7 (thin grey line) stained TIGR6B] in the expected manner (Fig. 1). Although Hyp6BM8 bound to all the strains shown in Fig. 1, its binding is selective to serogroup 6 as it did not stain a 9V strain (data not shown). These mAb binding patterns clearly show that the two isolates were serologically comparable to TIGR6X1 but distinct from serotypes 6A, 6B and 6C.
Fig. 1.
Flow cytometry profiles of various pneumococcal isolates stained with mAbs. The mAbs used were Hyp6AG1 (thin black line), Hyp6AM3 (dashed line), Hyp6BM8 (thick black line), and Hyp6BM7 (thin grey line). Studies with controls showed the staining threshold to be about 20 fluorescence units (data not shown).
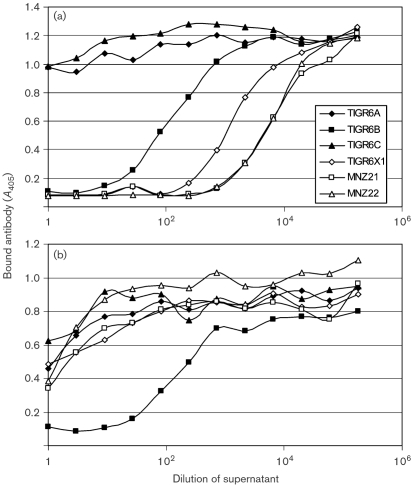
To further confirm that the two clinical isolates behave serologically as one would expect TIGR6X1 to behave, we examined these strains using an inhibition-type immunoassay which tests the ability of the PS to inhibit the binding of a mAb to 6B PS. Although Hyp6BM8 can bind to all members of serogroup 6 PS (Fig. 1), this mAb preferentially binds to 6B PS quite strongly and it was previously shown that TIGR6A and TIGR6C lysates did not inhibit its binding to 6B PS (Bratcher et al., 2009), although they are able to inhibit the binding of Hyp6BM8 to 6A PS (data not shown). We confirmed here again that TIGR6A and TIGR6C lysates did not inhibit the binding of Hyp6BM8 to 6B PS (Fig. 2) even though the lysates of TIGR6A and TIGR6C contained 6A and 6C capsular PS respectively (demonstrated by 6A and 6C PS assays; data not shown). In contrast, binding of Hyp6BM8 to 6B PS could be inhibited by TIGR6B lysate as well as by the lysates of the three strains TIGR6X1, MNZ21 and MNZ22 (Fig. 2a). This indicates that MNZ21 and MNZ22 do not belong to serotypes 6A and 6C. The two clinical isolates could be distinguished from serotype 6B with Hyp6BM7, which specifically reacts with 6B PS only. Its ability to bind to 6B PS could be inhibited with TIGR6B lysate, but not with the lysates of the three strains TIGR6X1, MNZ21 and MNZ22 (Fig. 2b). In addition, TIGR6X1, MNZ21 and MNZ22 PSs failed to inhibit the binding of Hyp6AG1 and Hyp6AM3 to 6A PS-coated plates (data not shown). These serological studies clearly show that MNZ21 and MNZ22 behave like TIGR6X1.
Fig. 2.
Binding of mAbs (y-axis) to 6B PS-coated ELISA plates in the presence of varying amounts of bacterial lysates (x-axis). The mAbs used were Hyp6BM8 (a) and Hyp6BM7 (b). The bacterial isolates studied are listed in the figure key.
Genetic findings
TIGR6X1 was created genetically by inserting the wciNβ gene into a 6B capsule gene locus (Bratcher et al., 2009). Thus, wciNβ and wciP6B are the two genetic features of 6X1. To investigate the presence of these two genetic features in isolates MNZ21 and MNZ22, we PCR amplified the wciN and wciP regions as we described before (Bratcher et al., 2009) and determined the nucleotide sequences of the amplicons. The sequences have been deposited in GenBank (accession no. GQ848645 for MNZ21 and GQ848646 for MNZ22). The sequences from the two isolates were identical, suggesting that they share a single origin. Their wciN sequences are strikingly different from the wciNα sequence (GenBank accession no. CR931638), but are highly homologous [99.6 % identity (1650/1656 bp) in the region sequenced using the PCR product from primers 5106 and 3101] to the wciNβ sequence of strain CHPA388 (Park et al., 2007a) in GenBank (accession no. EF538714). A characteristic of 6B wciP is the presence of an A at position 584 (based on the sequence of wciP in a previous publication: Mavroidi et al., 2004), which creates a codon for asparagine at residue 195 of the 6B WciP protein. In contrast, wciP from 6A or 6C has G at the corresponding position, creating a codon encoding serine (Mavroidi et al., 2004). The two new isolates had A at the corresponding position (GenBank accession no. GQ848645 and GQ848646). Thus, isolates MNZ21 and MNZ22 have the two genetic features of the 6X1 serotype.
In addition, we performed multi-locus sequence typing (MLST) analysis with MNZ21 and MNZ22 using the established method for S. pneumoniae (Enright & Spratt, 1998). The experiment showed that the two strains have identical sequences for the 3199 bp involved in MLST and both belong to ST282. As these isolates were collected from the same day-care centre in Jeju island in Korea, we suspect that these isolates are two members of the same clone.
Biochemical findings
Biochemical features distinguishing the PSs of the four members in the serogroup 6 are (a) rhamnose–ribitol linkage and (b) the presence of glucose instead of galactose. The 6A and 6B PSs contain galactose while 6X1 and 6C PSs do not. Capsular PSs from 6B and 6X1 have a 1→4 rhamnose–ribitol linkage, whereas the PSs from 6A and 6C have a 1→3 rhamnose–ribitol linkage. The two rhamnose–ribitol linkages can be readily distinguished because the 1→3 linkage is very sensitive to alkali hydrolysis, but the 1→4 linkage is not (Zon et al., 1982).
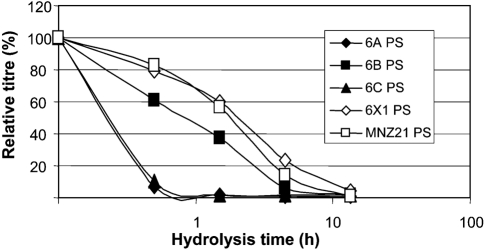
To distinguish between the two rhamnose–ribitol linkage types, we investigated capsular PS from MNZ21 to determine its susceptibility to alkali hydrolysis. As shown in Fig. 3, serotype 6A and 6C PSs retained less than 1 % of their original antigenicity after 1.5 h of alkali hydrolysis. In contrast, 6B, 6X1, and MNZ21 PSs retained significantly more of their antigenicity after 0.5 and 1.5 h. Thus, MNZ21 PS is as resistant to alkali hydrolysis as are the 6B and 6X1 PSs and therefore must not have the hydrolytically unstable 1→3 linkage.
Fig. 3.
Amount of antigenicity (y-axis) remaining after alkali hydrolysis for different time periods (x-axis). The PSs studied are listed in the figure key.
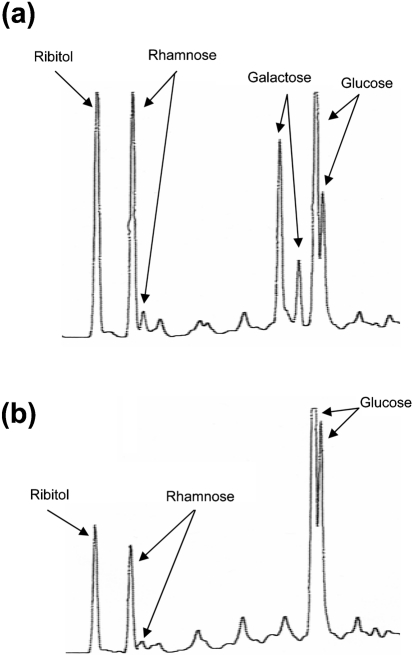
Studies of 6X1 PS showed that the product of wciNβ inserts glucose instead of galactose (Bratcher et al., 2009). To determine that the wciNβ gene in the natural isolates is functional, we purified capsular PS from MNZ21 and determined its component carbohydrates (Fig. 4b). When we analysed 6B PS for component carbohydrates, we found peaks for ribitol, rhamnose, galactose and glucose (marked in Fig. 4a). Also, the relative peak heights for the four carbohydrates were comparable. For instance, the major rhamnose peak was as tall as the dominant glucose peak. When we examined the component carbohydrates of MNZ21 PS (Fig. 4b), we found peaks for ribitol, rhamnose and glucose, but we did not find the galactose peaks which precede the glucose peaks in 6B PS. Also, in contrast to 6B PS (Fig. 4a), the major glucose peak of MNZ21 PS was much taller than the major rhamnose peak (Fig. 4b). This is also consistent with the conclusion that MNZ21 PS has one rhamnose but two glucose residues per repeating unit. Thus, the wciNβ gene in MNZ21 PS is functional, and MNZ21 capsular PS has glucose instead of galactose.
Fig. 4.
GLC profile of serotype 6B PS (a) and the PS purified from a pneumococcal isolate MNZ21 (b). The GLC instrument used here identified two peaks for rhamnose, two peaks for galactose and two peaks for glucose (Dr D. Pritchard, personal communication); these peaks are labelled in (a) along with the ribitol peak. The elution times (in minutes) for the labelled peaks (from left to right) are: (a) 10.325 (for ribitol), 10.692 and 10.805 (for rhamnose), 12.219 and 12.414 (for galactose), and 12.585 and 12.585 (for glucose); (b) 10.323, 10.690, 10.803, 12.582 and 12.655.
The above serological, genetic and biochemical studies clearly show that capsule type 6X1 can be expressed among natural pneumococcal isolates. Also, since the submission of our manuscript, pneumococcal isolates possessing the genetic characteristics of serotype 6X1 were discovered in Fijian islands (Jin et al., 2009). In view of these results, we suggest that the serotype designation 6D should be used in place of 6X1. Furthermore, finding natural isolates expressing the 6D capsule type in two very distant parts of the world suggests that this serotype is widespread throughout the world and that typing for 6D should be included in the serotyping scheme for pneumococci.
The discovery of serotype 6D completes the search for the potential members of serogroup 6, but it now raises new questions to be studied. For instance, the prevalence of the 6D serotype should now be investigated in different parts of the world, among different disease types, and among different age groups. As the use of pneumococcal vaccines may influence the prevalence of 6D as it did the prevalence of 6C (Nahm et al., 2009; Park et al., 2008), discovery of the 6D serotype will also have an important impact on future studies of pneumococcal conjugate vaccines.
Acknowledgments
We thank Dr D. Pritchard, S. Dong, J. Yu and I. Park for discussions and assistance. The work was supported with NIH funding (AI-31473) to M. H. N.
Abbreviations
PS, polysaccharide
Footnotes
References
- Avery, O. T. & Dubos, R. (1931). The protective action of a specific enzyme against type III pneumococcus infection in mice. J Exp Med 54, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, J. R., Liu, C., Dong, S. & Pritchard, D. G. (2006). Endopeptidase and glycosidase activities of the bacteriophage B30 lysin. Appl Environ Microbiol 72, 6825–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratcher, P. E., Park, I. H., Hollingshead, S. K. & Nahm, M. H. (2009). Production of a unique pneumococcal capsule serotype belonging to serogroup 6. Microbiology 155, 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, R. (1913). Treatment of pneumonia by means of specific serums. JAMA 61, 663–666. [Google Scholar]
- Enright, M. C. & Spratt, B. G. (1998). A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144, 3049–3060. [DOI] [PubMed] [Google Scholar]
- Fedson, D. S. (1988). Pneumococcal vaccine. In Vaccines, pp. 271–299. Edited by S. A. Plotkin & E. A. Mortimer, Jr. Philadelphia: W.B. Saunders.
- Jin, P., Kong, F., Xiao, M., Oftadeh, S., Zhou, F., Liu, C., Russell, F. & Gilbert, G. L. (2009). First report of putative Streptococcus pneumoniae serotype 6D among nasopharyngeal isolates from Fijian children. J Infect Dis 200, 1375–1380. [DOI] [PubMed] [Google Scholar]
- Kamerling, J. P. (2000). Pneumococcal polysaccharides: a chemical view. In Streptococcus Pneumoniae Molecular Biology and Mechanisms of Disease, pp. 81–114. Edited by A. Tomasz. Larchmont: Mary Ann Liebert.
- Lin, J., Kaltoft, M. S., Brandao, A. P., Echaniz-Aviles, G., Brandileone, M. C., Hollingshead, S. K., Benjamin, W. H. & Nahm, M. H. (2006). Validation of a multiplex pneumococcal serotyping assay with clinical samples. J Clin Microbiol 44, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroidi, A., Godoy, D., Aanensen, D. M., Robinson, D. A., Hollingshead, S. K. & Spratt, B. G. (2004). Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J Bacteriol 186, 8181–8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahm, M. H., Lin, J., Finkelstein, J. A. & Pelton, S. I. (2009). Increase in the prevalence of the newly discovered pneumococcal serotype 6C in the nasopharynx after introduction of pneumococcal conjugate vaccine. J Infect Dis 199, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I. H., Park, S., Hollingshead, S. K. & Nahm, M. H. (2007a). Genetic basis for the new pneumococcal serotype, 6C. Infect Immun 75, 4482–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I. H., Pritchard, D. G., Cartee, R., Brandao, A., Brandileone, M. C. & Nahm, M. H. (2007b). Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol 45, 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I. H., Moore, M. R., Treanor, J. J., Pelton, S. I., Pilishvili, T., Beall, B., Shelly, M. A., Mahon, B. E. & Nahm, M. H. (2008). Differential effects of pneumococcal vaccines against serotypes 6A and 6C. J Infect Dis 198, 1818–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, K. A., Baughman, W., Rothrock, G., Barrett, N. L., Pass, M., Lexau, C., Damaske, B., Stefonek, K., Barnes, B. & other authors (2001). Epidemiology of invasive Streptococcus pneumoniae infections in the United States, 1995–1998: opportunities for prevention in the conjugate vaccine era. JAMA 285, 1729–1735. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Hwang, Y. & Nahm, M. H. (2001). Avidity, potency, and cross-reactivity of monoclonal antibodies to pneumococcal capsular polysaccharide serotype 6B. Infect Immun 69, 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon, G., Szu, S. C., Egan, W., Robbins, J. D. & Robbins, J. B. (1982). Hydrolytic stability of pneumococcal group 6 (type 6A and 6B) capsular polysaccharides. Infect Immun 37, 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]