Abstract
Based on newly obtained 28S rDNA sequences from Roselliniella atlantica and R. euparmeliicola sp. nov., the genus Roselliniella has to be placed in Hypocreales and not in Sordariales; however, the family placement could not be resolved from the sequences obtained. The mature ascospores are single-celled and brown, but young ascospores are hyaline and sometimes have a median septum. The new species occurs on a Parmelia s.str. species in China, and differs in 24 nucleotide substitution positions in the nu-LSU rDNA region and ascospore size from R. atlantica. In this case, small variations in ascospore sizes and shape prove to be phylogenetically and taxonomically informative. The two species occur in the same clade with 95 % jack-knife support. Roselliniella atlantica occurs on Xanthoparmelia and Melanohalea species in Europe, whereas R. euparmeliicola was found on the species of Parmelia s.str. DNA was successfully recovered from a dried specimen of R. atlantica collected in 1992. Two unidentified fungi were also recovered from the Chinese specimen, and these belong to Sordariomycetidae and Dothideomycetes; whether these two are additional fungi living endolichenically in the lichen host, saprobes, or contaminants could not be ascertained.
Keywords: Ascomycota, endolichenic fungi, Hypocreales, lichenicolous fungi, Parmelia, Sordariales, Xanthoparmelia
INTRODUCTION
The generic name Roselliniella was introduced by Vainio (1921: 214) for pyrenocarpous fungi growing on lichens with single-celled brown ascospores and persistent filamentous interascal filaments. Vainio did not mention any included species, but referred to Saccardo (1882: 175) where three species were mentioned as comprising Rosellinia sect. Lichenicolae Sacc. 1882. The genus was generally treated as a later synonym of Adelococcus Theiss. & Syd. 1918, until resurrected first by Hafellner (1985). However, Hafellner based his concept of the genus on Endococcus haplospora Th. Fr. & Almq. 1867 which has thick-walled polyspored asci and paraphysoids, and this was not one of the original species mentioned by Saccardo. Santesson (in Eriksson & Hawksworth 1986: 310) drew attention to this misapplication, and introduced the new generic name Rosellinula for E. haplospora (syn. Muellerella thallophila Arnold 1888).
Santesson (in Eriksson & Hawksworth 1986) noted that lectotypification of Vainio’s generic name should await a revision of the genera involved. This was done by Matzer & Hafellner (1990: 53) who designated R. nephromatis (P. Crouan) Matzer & Hafellner 1990 as the type species, and recognised eight additional species, all growing on lichens, and mostly occurring in the Southern Hemisphere. Since that time, six further species have been added to the genus, and we describe an additional one as new here.
The systematic position of Roselliniella has not been resolved by molecular phylogenetic methods. The thin-walled functionally unitunicate I-asci, with no specialised apical apparatus, filamentous interascal filaments, and in some species the brown single-celled ascospores becoming delicately ornamented, led Hoffmann & Hafellner (2000: 91) to place the genus in the order Sordariales, but not to any family within the order. This classification has been followed in subsequent editions of the ‘Outline of Ascomycota’ (e.g. Eriksson 2006: 62, Lumbsch & Huhndorf 2007). However, it was not referred to an order, but only to the subclass Sordariomycetidae in Kirk et al. (2008), but that was only a lapsus that has now been changed in the database from which that was prepared (P.M. Kirk, pers. comm.).
The objectives of this study were to use molecular approaches to: 1) determine the placement of the genus in the Ascomycota more precisely; and 2) confirm that the modest differences in ascospore size between the new species and one already described from different parmelioid lichens were a valid taxonomic character.
MATERIALS AND METHODS
Specimens examined
In addition to the type collection of the new species, the following specimens of Roselliniella atlantica Matzer & Hafellner 1990 were studied for comparison.
Specimens examined. France, Campénéac, Morbihan, on Xanthoparmelia mougeotii on rock escarpment, 2 Apr. 1970, B.J. Coppins, E 00235005, paratype; Finistère, Chaos de St Herbot, on X. mougeotii, 8 June 1970, A.R. Pentecost, E 00235006, paratype. – United Kingdom, Outer Hebrides, Lewis, Port Geiraha, on X. mougeotii on rock, Aug. 1959, S.A. Manning, IMI 211909, paratype; Westerness, NE of Strontian, Ariundle Wood National Nature Reserve, on Melanohalea exasperata, 18 June 1992, A.M. O’Dare (Coppins 15370), E 00235007.
Morphological study
Specimens were studied using a Nikon zoom stereomicroscope and an Olympus BH-2 research microscope fitted with Nomarski differential interference contrast optics and a drawing tube. Freezing microtome sections were made on a Bright Starlet Cryostat, and both sections and squash preparations were used for microscopic examination. Preparations and measurements were made in water, lactofuchsin, and lactophenol cotton blue, the last two after warming.
Choice of additional taxa and outgroup
In addition to Roselliniella specimens we included representatives of other groups in Sordariomycetes and Dothideomycetes. Particularly, one species of Xylariales, five species of Dothideomycetes, 18 species of Hypocreomycetidae and 14 species of Sordariomycetidae, the two later respectively representing the four and six orders considered by Hibbett et al. (2007) for these two subclasses. Two specimens of Peziza (Pezizales, Pezizomycetes) were used as outgroup.
DNA extraction
DNA was extracted directly from dried specimens. Perithecia growing on the host thallus were carefully separated to avoid as much as possible obtaining tissue from the host with the point of a scalpel blade. Total DNA was extracted using the Qiagen DNeasy Plant MiniKit, according to the manufacturer’s instructions from perithecia of Roselliniella atlantica and R. euparmeliicola.
Amplification and sequencing
A fragment of c. 1 000 bp in the nuLSU was amplified using the primers LR0R (Vilgalys, www.biology.duke.edu/fungi/mycolab/primers.htm) and LR5 (Vilgalys & Hester 1990).
PCR amplifications were performed using Illustra™ Hot Start PCR beads, according to the manufacturer’s instructions, with the following settings: initial denaturalization of 95 °C for 3 min, four cycles (95 °C for 40 s, 56 °C for 40 s and 72 °C for 90 s), four cycles (95 °C for 30 s, 53 °C for 30 s and 72 °C for 90 s) and finally 32 cycles (95 °C for 30 s, 50 °C for 30 s and 72 °C for 90 s) with a final extension of 72 °C for 6 min.
When cloning was necessary, PCR products were ligated to a PCR 2.1-TOPO plasmid vector and cloned using TOPO TA Cloning kit according to the instructions of the manufacturer’s. Colonies were screened for the presence of the desired product, using the same primers and PCR programs described above. Clones with inserts of the appropriate size were sequenced.
Before sequencing, the PCR products were purified using the Viogene PCR-M Clean-up System or the enzymatic method Exo-sap-IT©.
Sequence alignment
Sequences were aligned using the software MAFFT v6.611 (Katoh et al. 2002, Katoh & Toh 2008) using the same procedures described in Wedin et al. (2009). This multiple alignment software has been frequently mentioned as one of the most accurate ones currently available (Niun et al. 2006, Carroll et al. 2007, Golubchik et al. 2007, Wedin et al. 2009). The ambiguous regions in the alignment were identified and eliminated using Gblocks v0.91b (Castresana 2000) with the following parameters: minimum number of sequences for a conserved position ‘20’, minimum number of sequences for a flank position ‘20’, maximum number of contiguous non-conserved positions ‘10’, minimum length of a block ‘5’ and allowed gap positions ‘with half’.
Parsimony and parsimony jack-knifing analyses
The analyses were performed using PAUP v4.0b10 (Swofford 2002) with the following settings: gaps are treated as ‘missing data’, 1 000 random addition sequence replicates, TBR branch swapping, steepest descent off, collapse branches if minimum length is 0, and MulTrees on. Jack-knife for identification of well-supported monophyletic groups (Farris et al. 1997) was performed in PAUP, with the following settings: heuristic search settings identical with the above analysis but with 20 random addition replicates; jack-knife settings: 1 000 jack-knife replicates with ‘JAC’-emulation, nominal deletion of characters 37 %, full heuristic search, retain groups with frequency > 50 %.
RESULTS
Taxonomy
Roselliniella euparmeliicolaMillanes & D. Hawksw., sp. nov. – MycoBank MB515195; Fig. 1, 2
Fig. 1.
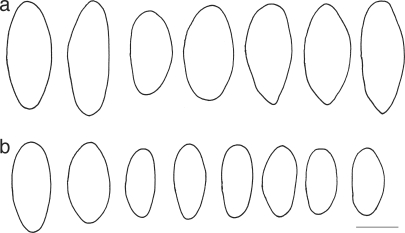
Ascospore outlines of Roselliniella species. a. R. atlantica (IMI 211909 paratype); b. R. euparmeliicola (BM holotype). – Scale bar = 10 μm.
Fig. 2.
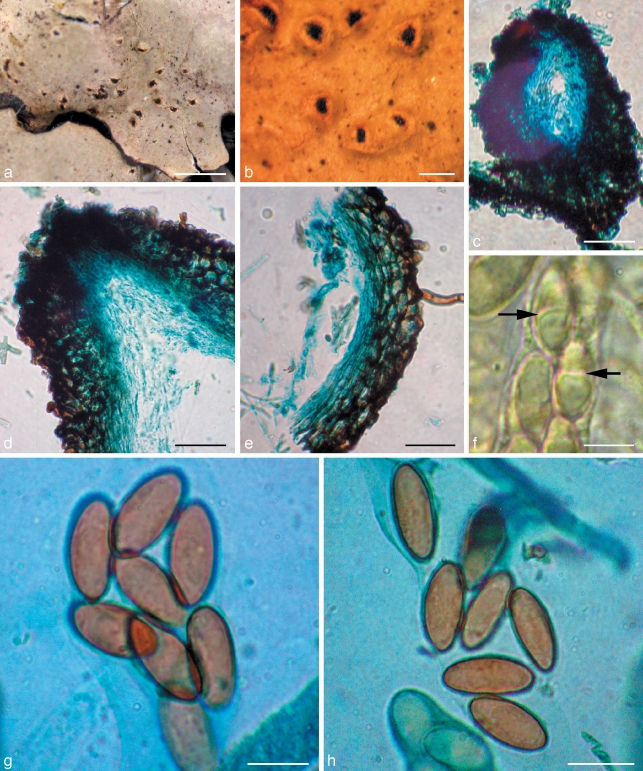
Roselliniella euparmeliicola (BM holotype). a. Infected thallus of Parmelia meiophora; b. urceolate warts with perithecia; c. vertical section of perithecium; d. vertical section of ostiolar region of perithecium; e. vertical section of perithecial wall; f. asci containing young ascospores, some with a medium septum (arrows); g, h. mature ascospores. — Scale bars: a = 2 mm; b = 250 μm; c = 100 μm; d, e = 50 μm; f–h = 10 μm.
Similis Roselliniella atlantica, sed ascosporiis minoribus, 17–20(–22) × 8–9.5(–12) μm.
Etymology. The species epithet indicates that the fungus lives on Parmelia s.str. species.
Ascomata perithecia, erumpent through the cortex of the host lichen, the cortex raised into wart-like convex to urceolate swellings, splitting apically or linearly or into teeth-like projections to expose the upper part of the perithecium, black, the area around the ostiole somewhat shiny, broadly pyriform, ostiole well-developed, (145–)170–410(–430) μm in surface view; exciple dark red-brown in section, intensifying in 10 % KOH, composed of 3–6 layers of pseudoparenchymatous cells, wall mainly 12.5–21 μm thick, but to 38 μm thick at the base and to 50 μm thick below the ostiole, with the innermost 5–7 μm layers of cells hyaline; cells irregularly polyhedral and somewhat angular, 7.5–10(–11) μm diam in surface view, smooth-walled, walls to 4–5 μm thick, radially compressed in vertical section and elongated vertically around the ostiole, cells in the innermost hyaline layers especially so; hyphae extending out from the lower parts and sides of the ascomata, pale brown to red-brown, flexuose, branched, smooth-walled, repeatedly septate and tending to be somewhat inflated between the septa, 3–4.5 μm thick. Hamathecium of periphyses and interascal filaments; periphyses abundant in the ostiolar canal, filamentous, septate, 1.5–2 μm thick; interascal filaments well-developed, persistent, filamentous, unbranched, septate, 1.5–2 μm thick; centrum I- (Lugol’s solution after pre-treatment in 10 % KOH). Asci elongate-clavate to subcylindrical, stalked, unitunicate in structure, walls ± equal in thickness and without any distinct apical thickening or internal apical structures, (52–)77–85(–95) × (12.5–)14–17 μm, 8-spored. Ascospores overlapping and irregularly biseriate in the ascus, non-septate, ellipsoid, rounded at the apices, lacking germ-pores, hyaline at first and occasionally with a median septum while still in the ascus, becoming pale to dark brown before release from the ascus, smooth-walled, the wall 0.5–1 μm thick,17–20(–22) × 8–9.5(–12) μm, length/width ratio 1.8–2.6 (av. = 2.1), with a thin hyaline perispore c. 1.5 μm thick, not or slightly swelling in 10 % KOH (excluded from the measurements).
Specimen examined. China, Yunnan Province, Jade Dragon Mountain, White River Stop, on Parmelia meiophora, 5 Oct. 2005, M.A. Allen & B.H. Hilton 405-8-2(c), holotype BM 000920346.
Host — In the thallus of Parmelia meiophora, apparently commensalistic as the infected areas of the thallus are not discoloured and there was no sign of necrosis.
Distribution — China (Yunnan Province); known only from the holotype collection.
Notes — This new species on Parmelia s.str. is very similar to Roselliniella atlantica Matzer & Hafellner 1990, a taxon first referred to by Hawksworth (1978: 181) as ‘Adelococcus cfr. groedensis (Zopf) Keissl. 1930’ on the basis of the similarly sized ascospores as reported in the literature. However, Matzer & Hafellner (1990) showed that A. groedensis had ascospores with apical pores, and when mature a septum near each apex, and occurred on Pertusaria lactea; they described the new genus Roselliniopsis Matzer & Hafellner 1990 to accommodate that fungus and a related tropical species.
The ascospores in the specimens of Roselliniella atlantica we examined for comparison in this study were (18–)22–26(–29) × (11.5–)12–14(–14.5) μm, thus being longer and broader than in the new species (Fig. 1); the shortest (from 17 μm) and narrowest (from 9 μm) ends of the range measurements in Hawksworth (1978) were based on somewhat immature spores. Also, according to Ihlen & Wedin (2008: 315) they can reach 30 μm in length. Matzer & Hafellner (1990: 57, fig. 5c) illustrate one spore with a septum in the lower third. We saw no septa in the slides we made of this species, either in hyaline immature or brown mature spores. In addition to the differences in size, the ascospores of R. euparmeliicola tend to be somewhat more pointed at one or both ends.
Roselliniella atlantica occurs on two species of a different parmelioid lichen genus from the new species: Xanthoparmelia (incl. Neofuscelia), notably the yellow-green X. mougeotii, and the brown X. verruculifera, and is also reported here on Melanohalea exasperata. That fungus is only known from Europe, where it occurs in France, Sweden, and the UK. In the specimen IMI 211909, it may be significant that the perithecia were not developed on two other parmelioid lichens growing mixed with X. mougeotii on the piece of rock: Melanelixia fuliginosa and Parmelia omphalodes. The specimen on Melanohalea exasperata, with ascospores we measured as 23.5–26.5 × (10.5–)12–13 μm, agrees with R. atlantica in all the features we could study on the rather sparse material. The ascomata of R. atlantica also differ from those of the new species in the way they originate on the different host lichens, emerging erumpently from the thallus but without producing raised wart-like structures with splits where the cortex has been forced apart. The species may also be more pathogenic than R. euparmeliicola as infected lobes of X. mougeotii become discoloured and friable, so that infected scraps are often all that is found.
Phylogenetic analysis
We generated five new sequences of the 28S rDNA (Table 1) which were aligned together with sequences already available in GenBank. Three of the five new sequences which were amplified during cloning experiments corresponded to other fungi in the sample, and were different from those of the host genus Parmelia s.str.
Table 1.
Sequences newly produced (bold) or downloaded from GenBank. Specimen numbers are given for newly produced sequences.
| Taxon | Specimen | GenBank accesion number (n-LSU) |
|---|---|---|
| Bionectria ochroleuca | AY686634 | |
| Bionectria pityrodes | AY489728 | |
| Botryosphaeria ribis | AY004336 | |
| Camarops petersii | AY346265 | |
| Chaetosphaerella phaeostroma | AY346274 | |
| Chaetosphaeria lateriphiala | AF466072 | |
| Chaetosphaeria pygmaea | AF466077 | |
| Coniochaeta sp. | AY346275 | |
| Coniochaetidium savoryi | AY346276 | |
| Coniothyrium obiones | DQ678054 | |
| Cordyceps tuberculata | AF327384 | |
| Cordyceps militaris | AF327374 | |
| Cosmospora coccinea | AY489734 | |
| Cosmospora vilior | AY015626 | |
| Cylindrocarpon lichenicola | AY097324 | |
| Diaporthe phaseolorum | AY346279 | |
| Hypocrea rufa | AY489726 | |
| Lasiosphaeriella nitida | AY346289 | |
| Leptosphaeria maculans | AY849946 | |
| Leptosporella gregaria | AY346290 | |
| Melanospora singaporensis | AY015629 | |
| Melanospora zamiae | AY046579 | |
| Microascus trigonosporus | DQ470958 | |
| Mycopepon smithii | AF279400 | |
| Nectriopsis sporangiicola | AF210661 | |
| Neonectria fuckeliana | AY283551 | |
| Neonectria radicicola | U17415 | |
| Neurospora crassa | AF286411 | |
| Nitschkia grevillei | AY346294 | |
| Ophiostoma piceae | AF234837 | |
| Peziza proteana | AY544659 | |
| Peziza vesiculosa | AY500552 | |
| Poroconiochaeta discoidea | AY346297 | |
| Roselliniella atlantica | E-235007 | GQ888763 |
| Roselliniella euparmeliicola | BM-920346 | GQ888764 |
| Schizoparme botrytidis | AF408383 | |
| Sordaria macrospora | AY346301 | |
| Sphaerodes fimicola | AY015628 | |
| Unknown Dothideomycetes-a | BM-920346 | GQ888765 |
| Unknown Dothideomycetes-b | BM-920346 | GQ888766 |
| Unknown Sordariomycetidae | BM-920346 | GQ888767 |
| Valsa ceratosperma | AF408387 | |
| Vittatispora coorgii | DQ017375 | |
| Xylaria hypoxylon | AY544648 |
The two Roselliniella sequences differed in 24 nucleotide positions, and formed a single clade with 95 % jack-knife support.
The matrix analysed contained 1 057 aligned characters. After the exclusion of ambiguously aligned and uninformative sites, 376 parsimony informative sites were used in the analyses. Our parsimony analysis resulted in six equally most parsimonious trees (1 739 steps, CI = 0.389, RI = 0.657 and RC = 0.256).
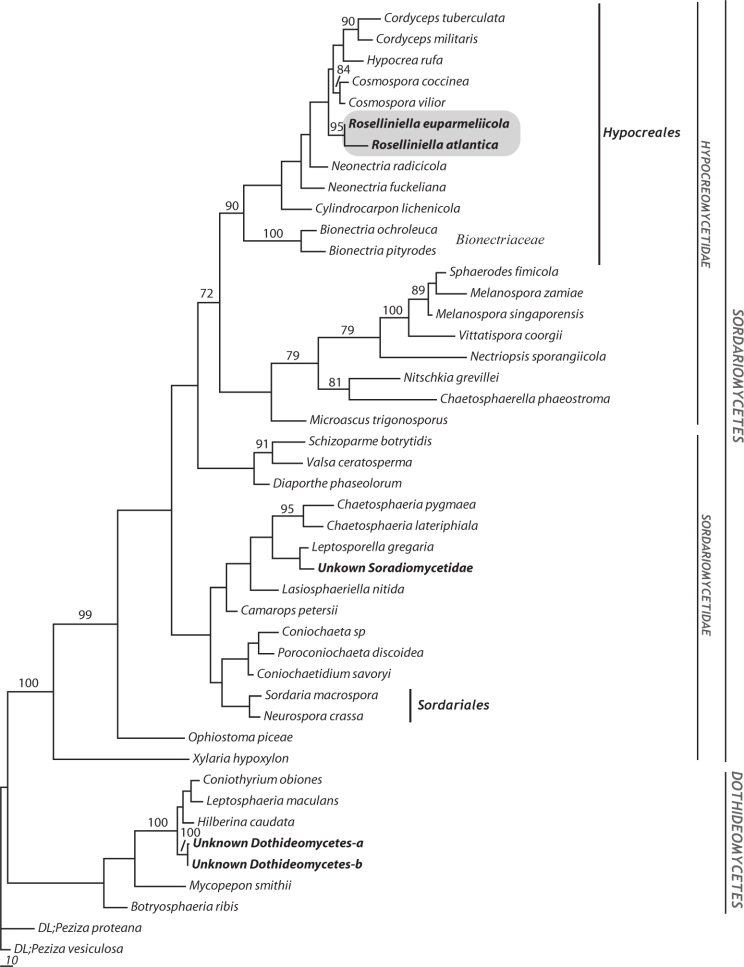
The species in Hypocreales we included in the analysis formed a monophyletic clade with 89 % jack-knife support, and both Roselliniella species were included within it. The two Roselliniella species joined with 95 % jack-knife support and were a sister group to a clade including species in Nectriaceae (Fig. 3), though without significant support.
Fig. 3.
One of the 6 most parsimonious trees based on the analysis of 376 informative sites of 44 nucLSU sequences; length 1 739, CI = 0.389, RI = 0.657 and RC = 0.256. Support measured by jack-knifing (≥ 70 %) is represented with the numerical values written above the corresponding branches. Branch lengths are proportional to the number of nucleotide differences. Sequences new generated are in bold, and a shaded box states the clade including both species of Roselliniella sequenced in this study. Selected class, subclass, order and family designations following Hibbet et al. (2007) are given in the right side. Two specimens of Peziza (Pezizales, Pezizomycetes) were used as outgroup.
Sequences of three additional fungal taxa were isolated from the Parmelia meiophora thalli during cloning experiments. One of them is placed within the Chaetosphaeriales clade, whereas the remaining two group with Dothideomycetes representatives in a strongly supported clade (100 % jack-knife value). None had matches in GenBank and remain unidentified.
DISCUSSION
In our phylogenetic analyses, the two species of Roselliniella were conclusively placed in the Hypocreales clade, though the family placement within the order was not recovered with phylogenetic support. There can consequently be no doubt that Roselliniella belongs to the order Hypocreales in class Sordariomycetes. This was a somewhat surprising result as the ascomata of Roselliniella species are erumpent and not superficial on a stroma, appear dark walled and have persistent interascal filaments, the asci lack an apical ring, and at maturity the ascospores are brown and single-celled rather than colourless or pale two- or more-celled. However, while ascospores that are single-celled from the first are known in some Hypocreales, for instance Nigrosabulum globosum in Bionectriaceae (Plishka et al. 2009), we found that immature hyaline spores of R. euparmeliicola, while inside the ascus, sometimes had a medium septum (Fig. 2f). The ascomatal walls are rather robust and dark coloured for Hypocreales, but the layered wall, with the cells somewhat swollen and rounded in surface view, does recall many members of the order. More fundamentally, while interascal filaments are characteristically absent in Hypocreales, ‘apical paraphyses’ resembling those of the Roselliniella species are described and illustrated in the type species of Nectria, N. cinnabarina, and also in N. lamyi, by Rossman et al. (1999: 143, pl. 30b, h). We were unable to determine the origins of the filaments in the Chinese specimen, and so could not resolve whether they were ‘apical’ or ‘true’ paraphyses; if the latter, thus would be most unusual for the family and merit an emendation of its features.
Biologically, it is interesting to see that Roselliniella appears in the hypocrealean clade that includes many fungicolous and also some lichenicolous fungi. Cosmospora species in particular mainly occur on perithecioid ascomycetes, though a few are entomogenous or found directly on plant material (Rossman et al. 1999). However, most lichenicolous hypocrealean genera (e.g. Nectriopsis, Paranectria, Pronectria, Trichonectria), are placed in Bionectriaceae, although some are in Nectriaceae (e.g. Xenonectriella). Cylindrocarpon lichenicola (syn. Fusarium lichenicola) was also placed in Hypocreales out of the Bionectriaceae clade in our tree, but we stress that this is not obligately lichenicolous but a primarily saprobic fungus known from diverse habitats (Hawksworth 1979).
In our first comparison of sequences with those in GenBank, Melanospora fallax U17404 and M. zamiae U17405 were in a clade with 100 % jack-knife support, and a sister group (but without support) to the two Roselliniella species. However, the LSU sequence from U17405 (Rehner & Samuels 1995) was found to be erroneous when the same isolate from the American Type Culture Collection (ATCC) was re-sequenced by Zhang & Blackwell (2002), and this must also be so for U17404 which those authors did not examine. Neither of these two sequences fall in the well-supported Melanosporales clade along with other species of Melanospora, Sphaerodes and Vittatispora (Chaudhary et al. 2006).
The extent to which small differences in ascospore sizes and shapes is phylogenetically informative in lichenicolous fungi is uncertain, especially where closely related hosts are involved as in the parmelioid lichens (Doré et al. 2006). In the case of the two Roselliniella species we investigated, the differences in 24 nucleotide positions show that R. atlantica and R. euparmeliicola represent different phylogenetic species, and consequently the differences in ascospore size ranges are informative in this case. However, we recognise that further studies including more Roselliniella species are needed to test the phylogenetic value of ascospore size as a synapomorphic character in the genus as a whole. Further, this result should not be automatically extrapolated to other cases in which small differences in ascospore size in lichenicolous fungi occurring on different host lichens are found. It does show, however, that in such instances molecular data should be obtained to see whether or not the variation is phylogenetically informative.
That the two Roselliniella species distinguished here occur on different genera of parmelioid lichens is of special interest as it appears to provide another instance where lichenicolous fungi per se are taxonomically and phylogenetically informative in this group of lichens. However, such conclusions must remain tentative when few specimens have been available for study. Correlations between lichenicolous fungi and the genera now recognized in Parmelia s.l. (Blanco et al. 2006) have previously been demonstrated in, for example, species of Abrothallus (Ihlen & Wedin 2008) and Homostegia (Hawksworth et al. 2004).
It is not possible to state whether the unidentified Chaetosphaeriales and Dothideomycetes fungi from the Chinese specimen are contaminants or asymptomatic taxa growing inside the host lichen, i.e. endolichenic fungi. It does appear that fungi other than that making the lichen thallus can commonly be detected by isolation into culture (Petrini et al. 1990, Möller & Dreyfuss 1996, Suryanarayan et al. 2005, Li et al. 2007) or molecular sequencing (Miadlikowska et al. 2005). What is not resolved is whether these fungi are actually living in the lichen tissues or if they are present as captured spores fortuitously present.
Our success in obtaining sequence data from a dried specimen of Roselliniella atlantica collected 15 years earlier, however, demonstrates that molecular approaches to studies of phylogenetic relationships and species concepts amongst lichenicolous fungi can be feasible in the absence of very recently collected material. We believe that this is the oldest material of a lichenicolous ascomycete to have sequence data obtained from it to date.
Acknowledgments
We are indebted to Margaret A. Allen and Barbara H. Hilton for allowing us to study the collection they made in China. Also, Brian J. Coppins (Edinburgh) and Milena Ross (Egham) are thanked for arranging the loan of specimens for comparison and sequencing. This investigation was undertaken as a part of a research grant to D.L.H. from the Ministerio de Educación y Ciencia of Spain (Proyectos I+D CGL 2008-01600) and supported by a grant from the Swedish Research Council (VR 621-2006-3760) to M.W.
REFERENCES
- Blanco O, Crespo A, Ree RH, Lumbsch HT. 2006. Major clades of parmelioid lichens (Parmeliaceae, Ascomycota) and the evolution of their morphological and chemical diversity. Molecular Phylogenetics and Evolution 39: 52 – 69 . [DOI] [PubMed] [Google Scholar]
- Carroll H, Beckstead W, O’Connor T, Ebbert M, Clement M, Snell Q, McClellan D. 2007. DNA reference alignment benchmarks based on tertiary structure of encoded proteins. Bioinformatics 23: 2648 – 2649 . [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540 – 552 . [DOI] [PubMed] [Google Scholar]
- Chaudhary P, Campbell J, Hawksworth DL, Sastry KN. 2006. Vittatispora, a new melanosporaceous genus from Indian soil. Mycologia 98: 460 – 467 . [DOI] [PubMed] [Google Scholar]
- Doré CJ, Cole MS, Hawksworth DL. 2006. Preliminary statistical studies of the infraspecific variation in the ascospores of Nesolechia oxyspora growing on different genera of parmelioid lichens. Lichenologist 38: 425 – 434 . [Google Scholar]
- Eriksson OE. (ed). 2006. Outline of Ascomycota 2006. Myconet 12: 1 – 82 . [Google Scholar]
- Eriksson OE, Hawksworth DL. 1986. Outline of the Ascomycetes 1986. Systema Ascomycetum 5: 185 – 324 . [Google Scholar]
- Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG. 1997. Parsimony jackknifing outperforms neighbor-joining. Cladistics 12: 99 – 124 . [DOI] [PubMed] [Google Scholar]
- Golubchick T, Wise MJ, Easteal S, Jermiin LS. 2007. Mind the gaps: evidence of bias in estimates of multiple sequence alignment. Molecular Biology and Evolution 24: 2433 – 2442 . [DOI] [PubMed] [Google Scholar]
- Hafellner J. 1985. Studien über lichenicole Pilze und Flechten III. Die Gattungen Roselliniella Vainio emend. Haf. (Ascomycotina, Dothideales). Herzogia 7: 145 – 162 . [Google Scholar]
- Hawksworth DL. 1978. Notes on British lichenicolous fungi: II. Notes from the Royal Botanic Garden Edinburgh 36: 181 – 197 . [Google Scholar]
- Hawksworth DL. 1979. The lichenicolous hyphomycetes. Bulletin of the British Museum (Natural History), Botany 6: 183 – 300 . [Google Scholar]
- Hawksworth DL, Atienza V, Cole MS. 2004. Lichenicolous species of Homostegia (Dothideomycetes), with the description of H. hertelii sp. nov., a new fungus on Flavoparemlia species. Bibliotheca Lichenologica 88: 187 – 194 . [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF , et al. 2007. A higher level phylogenetic classification of the Fungi. Mycological Research 111: 509 – 547 . [DOI] [PubMed] [Google Scholar]
- Hoffmann N, Hafellner J. 2000. Eine Revision der lichenicolen Arten der Sammelgattungen Guignardia und Physalospora (Ascomycotina). Bibliotheca Lichenologica 77: 1 – 181 . [Google Scholar]
- Ihlen PG, Wedin M. 2008. An annotated key to the lichenicolous Ascomycota (including mitosporic morphs) of Sweden. Nova Hedwigia 86: 275 – 365 . [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on Fast Fourier transform. Nucleic Acids Research 30: 3059 – 3066 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286 – 298 . [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008. Ainsworth & Bisby’s Dictionary of the Fungi 10th edn Wallingford, CAB International; . [Google Scholar]
- Li W-C, Zhou J, Guo S-Y, Guo L-D. 2007. Endophytic fungi associated with lichens in Baihua mountain of Beijing, China. Fungal Diversity 25: 69 – 80 . [Google Scholar]
- Lumbsch HT, Huhndorf SM. 2007. Outline of Ascomycota 2007. Myconet 13: 1 – 58 . [Google Scholar]
- Matzer M, Hafellner J. 1990. Eine Revision der lichenicolen Arten der Sammelgattung Rosellinia (ascomycetes). Bibliotheca Lichenologica 37: 1 – 138 . [Google Scholar]
- Miadlikowska J, Arnold AE, Higgins KL, Sarvate S, Gugger P, Way A, Hofstetter V, Lutzoni F. 2005. Endolichenic fungi: random inhabitants or symbiotic partners. Proceedings of the Annual Meeting of the Mycological Society of Japan 49: 162 . [Google Scholar]
- Möller C, Dreyfuss MM. 1996. Microfungi from Antarctic lichens, mosses and vascular plants. Mycologia 88: 922 – 933 . [Google Scholar]
- Nuin PAS, Wang Z, Tillier ERM. 2006. The accuracy of several multiple sequence alignment programs for proteins. BMC Bioinformatics 7: paper 471 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini O, Hake U, Dreyfuss MM. 1990. An analysis of fungal communities isolated from fruticose lichens. Mycologia 82: 444 – 451 . [Google Scholar]
- Plishka MJR, Tsuneda A, Currah RS. 2009. Morphology and development of Nigrosabulum globosum, a cleistothecial coprophile of the Bionectriaceae (Hypocreales). Mycological Research 113: 815 – 821 . [DOI] [PubMed] [Google Scholar]
- Rehner SA, Samuels GJ. 1995. Molecular systematics of the Hypocreales: a teleomorph gene phylogeny and the status of their anamorphs. Canadian Journal of Botany 73: S816 – S823 . [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, Lowen R. 1999. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1 – 248 . [Google Scholar]
- Saccardo PA. 1882. Sylloge Fungorum. Vol. 1 PA Saccardo, Padova: . [Google Scholar]
- Suryanarayan TS, Thirunavukkarasu N, Hariharan GN, Balayi P. 2005. Occurrence of non-obligate microfungi inside lichen thalli. Sydowia 25: 120 – 130 . [Google Scholar]
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10 Sinauer Associates, Sunderland, MA: . [Google Scholar]
- Vainio EA. 1921. Lichenographia fennica I. Acta Societatis Fauna Flora fennica 49, 2: 1 – 274 . [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238 – 4246 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedin M, Wiklund E, Jørgensen PM, Ekman S. 2009. Slippery when wet: phylogeny and character evolution in the gelatinous cyanobacterial lichens (Peltigerales, Ascomycetes). Molecular Phylogenetics and Evolution: doi:10.1016/j.ympev.2009.08.013 . [DOI] [PubMed] [Google Scholar]
- Zhang N, Blackwell M. 2002. Molecular phylogeny of Melanospora and similar pyrenomycetes fungi. Mycological Research 106: 148 – 155 . [Google Scholar]