Abstract
Recombinant tissue plasminogen (rt-PA) with 35 cysteine residues has been completely assigned by mapping the 17 disulfide linkages and the unpaired cysteine. The result is consistent with the prediction from homology except for the unassigned cysteine, which was identified at Cys83. This cysteine was found to be blocked and paired with either a glutathione or cysteine residue in a ~ 60 :40 ratio, respectively. The analysis was conducted using a multi-fragmentation approach consisting of ETD and CID, in combination with a multi-enzyme digestion strategy (Lys-C, trypsin, and Glu-C). The disulfide-linked peptides, even those containing N or O-linked glycosylation, could be assigned since the disulfide bonds were still preferably cleaved over the glycosidic cleavages under ETD fragmentation. The use of a multiple and sequential enzymatic digestion strategy was important in producing fragment sizes suitable for analysis. For the analysis of complex intertwined disulfides, the use of CID MS3 to target partially disulfide dissociated peptides from the ETD fragmentation was necessary for linkage assignment. The ability to identify the exact location and status of the unpaired cysteine (free or blocked with a glutathione or cysteine) could shed light on the activation of rt-PA, upon stimulation by either oxidative or ischemic stress.
Introduction
The comprehensive characterization of protein therapeutics or protein targets is a continual challenge, given the complexity of these biopolymers.1-5 In addition to post-translational modifications, disulfide linkages and the location and status of unpaired cysteines (free or blocked) are critical structural features that need to be determined. Disulfide linkages are a controlling factor in the three dimensional structure of proteins and are thus intimately involved in protein folding.6 The free cysteines can be reactive (most often in redox reactions), leading to covalent association7, enzyme catalysis8 or, alternatively, disulfide scrambling.9 It is also well known that unpaired cysteines on a protein may be blocked by the redox “buffer” glutathione, prior to secretion.10 In some cases, the reversible reaction of glutathione with cysteine or other oxidizing species can be a mechanism for pathway signaling.11 Thus, it is clear that disulfide linkage and unpaired cysteine location and status are important structural features that need to be determined as part of the comprehensive characterization of proteins for structure and function correlation.
The need to analyze disulfide linkages and the status of unpaired cysteines have led to the introduction of a variety of methods, such as Edman degradation12-13 and mass spectrometry using collision induced dissociation (CID) in either the positive or negative ion modes.14-16 With respect to the analysis of unpaired cysteines, the use of the Ellman reagent to quantitate the free cysteines with tagged fluorophores is often employed.17 However, the determination of intertwined disulfide linkages and the location of cysteines, either free or possibly bonded with other molecules, are still difficult to accomplish.
Disulfide bond cleavage has been shown to be favored over peptide backbone breakage for electron capture dissociation (ECD)19 and more recently for electron transfer dissociation (ETD).18 This preferred cleavage is likely due to the fact that free electrons can be more easily captured by sulfur (disulfides) than by backbone (amides) during the electron capture or transfer process.20 Recently, we have successfully mapped disulfide linkages using on-line LC-MS with ETD based on the favorable breakage of the disulfide bond.21 The cleaved or partially cleaved disulfides were further fragmented by CID-MS3 to obtain specific linkage locations.
In this work, we use multiple and sequential enzymatic digestion in conjunction with ETD to study the complex glycoprotein, recombinant tissue plasminogen activator (rt-PA) which has 35 cysteine residues (17 disulfides plus one potential free cysteine). Up to now, the disulfide assignments have been based on homology, and no direct evidence has been presented for the complete structure. Using this LC/MS with ETD approach, not only are all intertwined disulfide linkages determined, but the location of the unpaired cysteine is identified and found to be blocked either by glutathione or an additional cysteine.
Materials and Methods
Materials
Achromobacter protease I (Lys-C) was obtained from Wako Co. (Richmond, VA), Glu-C from Roche (Indianapolis, IN), and mass spectrometric grade trypsin from Promega (Madison, WI). Fluoranthene, guanidine hydrochloride and ammonium bicarbonate were purchased from Sigma-Aldrich (St. Louis, MO). Recombinant tissue plasminogen activator (Activase), consisting of 50 mg of the active protein was the gift of Genentech Inc (South San Francisco, CA). Additionally, a genetically engineered t-PA mutant (TNK), which has the same amino acid sequence as the wild type except with substitutions at 3 locations,T103N, N117Q, and KHRR (296-299) to AAAA22, was also kindly provided by Genentech. Formic acid and acetonitrile were purchased from Thermo Fisher Scientific (Fair Lawn, NJ), and HPLC-grade water, used in all experiments, was from J.T. Baker (Bedford, MA). Microcon YM-10 Centrifugal Filter Unit was obtained from Millipore (Bedford, MA).
Enzymatic digestion (without reduction and alkylation)
Protein solution (1 μg/μL) was buffer exchanged with 0.1 M ammonium bicarbonate (pH 8) over a Microcon spin column (10 kDa MWCO, Millipore). For Lys-C plus trypsin digestion, the protein solution (after buffer exchange) was added with endoproteinase Lys-C (1:50 w/w) for 8 hr at room temperature. Then, trypsin (1:50 w/w) was added to an aliquot from the Lys-C digestion for further reaction for 12 hr at room temperature, with the pH adjusted to 6.8. In order to minimize potential disulfide scrambling, a slightly less than alkaline pH was used in the enzymatic digestion protocol. For Lys-C plus trypsin, followed by Glu-C digestion, Glu-C (1:50 w/w) was added to an aliquot from the Lys-C plus trypsin digestion for an additional 8 hrs at room temperature, with the pH adjusted to 5.8. In all cases, digestion was terminated by addition of 1% formic acid.
LC-MS
LC-MS experiments were performed on an LTQXL with ETD mass spectrometer (Thermo Fisher Scientific, San Jose, CA), consisting of a linear ion trap with an additional chemical ionization source to generate fluoranthene anions. An Ultimate 3000 nanoLC pump (Dionex, Mountain View, CA) and a self-packed C8 column (Vydac C8, 300Å pore and 5 μm particle size, 75 μm i.d. × 10 cm) were coupled on-line to the mass spectrometer through a nanospray ion source (New Objective, Woburn, MA). Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1 % formic acid in acetonitrile. The gradient consisted of: (i) 20 minutes at 0 % B for sample loading; (ii) linear from 0 to 40% B over 40 min; (iii) linear from 40 to 80% B over 10 min; and finally (iv) isocratic at 80% B for 10 min. The flow rate of the column was maintained at 200 nL/min. In an analogous manner to our recent publication,21 the mass spectrometer was operated in the data-dependent mode to switch automatically between MS (scan 1), CID-MS2 (scan 2), ETD-MS2 (scan 3), and CID of isolated species in the MS3 steps (scan 4). The normalized collision energy used in CID (scans 2 and 4) were set at 35%. Briefly, after a precursor ion scan, the CID-MS2 and ETD-MS2 activation steps were performed on the same precursor ion. Each precursor ion was isolated using the data dependent acquisition mode with a ± 2.5 m/z isolation width to select automatically and sequentially a specific ion (starting with the most intense ion) from the first MS scan. Finally, an additional MS3 step, which isolated the highest intensity ion (or targeted ion) from the prior ETD spectrum for further CID fragmentation at 35% normalized collision energy (± 5 m/z isolation width) was implemented. To further confirm the peptide masses and charge states, an LTQ-FT MS (Thermo Fisher Scientific) was employed, as necessary, to acquire full mass spectra in the FT-ICR (400 2000 m/z) at 100,000 mass resolution to determine the accurate mass and charge states of the precursor ions generated under similar conditions to those in the LTQXL-ETD MS instrument. If two or more precursor ions with close m/z values appeared at overlapped retention times, the CID-MS2 spectrum pattern was then added to track the correct m/z precursor ion between the LTQ-ETD and LTQ-FT MS runs.
Results and Discussion
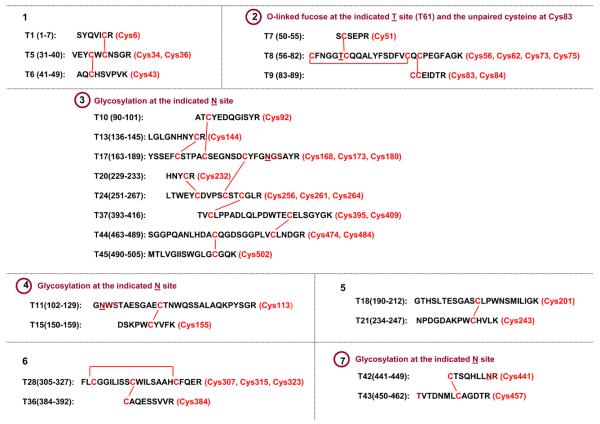
Recombinant tissue plasminogen activator (rt-PA) has 17 disulfide bonds and one unpaired cysteine, with linkages assigned based on sequence homology with other related protein families (e.g. epidermal growth factor and serine protease).22-23 When rt-PA is digested by trypsin (without disulfide reduction), the disulfide bonds are distributed in 7 tryptic peptides, with 4 out of the 7 peptides being glycosylated (3 N-linked and 1 O-linked), as shown in Figure 1. The three non-glycosylated peptides have been successfully analyzed in our previous paper using the ETD/CID strategy.21 The remaining four peptides with glycosylation are examined in this study. Initially, Lys-C plus trypsin digestion was employed to improve the digest of rt-PA.21 Additionally, Lys-C plus trypsin, followed by Glu-C, was used to further trim down the size of the peptides with multiple disulfides (e.g. peptides #2 and #3 in Figure 1) to produce sufficient fragmentation cleavages for assignment, as discussed in the following.
Figure 1.
Theoretical non-reduced tryptic peptides of rt-PA, including linked disulfides (lines), cysteine position (parenthesis), the peptides with glycosylation (cycles), and glycosylation sites (underlines).
Identification of peptide #2 with O-linked fucose and unpaired cysteine
As indicated in Figure 1, peptide #2 contains the unpaired cysteine along with an O-linked fucose. There has been a significant effort to pinpoint the exact position of the unpaired cysteine which is adjacent to a second cysteine, as well as to determine the status of this cysteine (i.e. whether the thiol is free or blocked). To date, no direct evidence has been presented on the location and state of the unpaired cysteine. This is likely due in part to the attached O-linked fucose, which results in mainly neutral loss by CID-MS2. In previous work24, we found that the fucose was still attached to the peptide backbone after ETD fragmentation, and this result allowed us to identify the fucose linkage position on the peptide (after the protein was reduced). In the present study with the disulfide intact, we further analyzed the peptide which contained the unpaired cysteine.
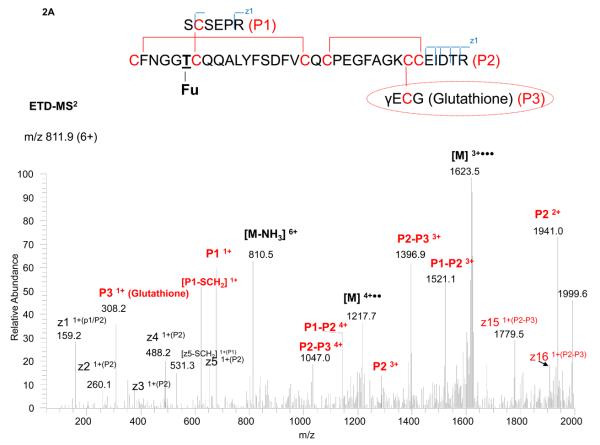
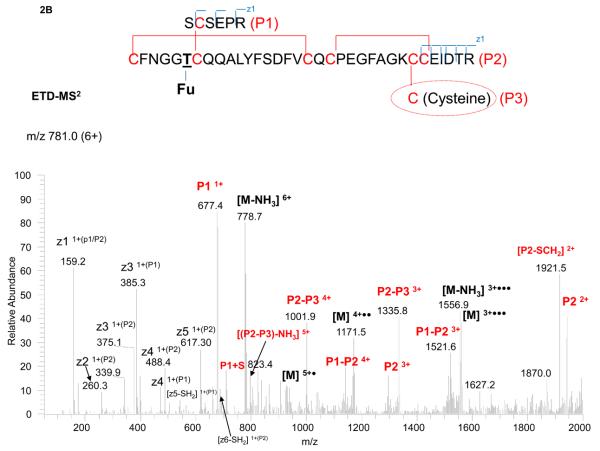
Interestingly, we could not find the expected mass of the peptide, assuming the unpaired cysteine was free. Instead, two other masses were found, and both were closely related to each other, based on their fragmentation patterns and difference in precursor masses. One precursor could be matched to the peptide paired with a glutathione and the other with a cysteine residue, as shown in Figures 2A and 2B. It should be noted the free cysteine, once forming a disulfide (i.e. paired with a glutathione or cysteine), prevents the enzyme (either Lys-C or trypsin) to cleave the amino acid “K” next to the disulfide. (In Figure 1, the assumption was made that the unpaired cysteine was free, and thus the lysine was cleaved.) Without digestion of this K, 18 Da (loss of water) must be subtracted from the resulting peptide mass to account for the miscleavage, as shown in Figures 2A (for glutathione blockage) and 2B (for cysteine blockage).
Figure 2.
ETD-MS2 spectrum of the disulfide-linked tryptic peptide with an O-linked fucose and unassigned cysteine paired with either a glutathione (2A) or cysteine (2B), with the peptide structure and annotation of the fragment ions indicated in the insert.
The precursor mass of 811.9 (6+) (for the peptide paired with a glutathione residue and with one miscleavage) was analyzed by ETD-MS2 (Figure 2A). The glutathione residue (P3 ion) was readily observed, along with ions for P1, P2, P1-P2 and P2-P3. The partially disulfide-dissociated peptides (P1-P2 and P2-P3) allowed direct assignment of the linked peptides as P1 associated with P2, and P2 with P3, but not P1 with P3. The exact linkages within P2 (i.e. Cys56 with Cys73, and Cys75 with Cys84) typically require CID-MS3 to further target the peptide ion for analysis.21 However, since the fucose was also located on P2, the additional CID-MS3 step produced, as expected, mainly the neutral loss of the fucose from the P2 ion. Since the P2 peptide ion, after the MS3 step, was still large (due to the miscleavage), the use of CID-MS4 to fragment the P2 ion further generated limited cleavages that were insufficient for structure assignment. As a result, we employed an additional Glu-C digestion step to cut the disulfide-linked peptide into smaller fragments. With these smaller fragments, even taking into account the neutral loss of fucose, the limited backbone cleavage for CID-MS2 provided sufficient information for structure assignment (see Figure S1 in the Supplementary Material). From the fragmentation observed, we were able to locate the unpaired cysteine to Cys83 and to determine that it was paired with a glutathione residue.
The other precursor mass of 781.0 (6+), for the peptide paired with a cysteine residue, was assigned in a similar manner. In this case, the cysteine residue (P3 ion) was too small to be observed in the ETD-MS2 spectrum (Figure 2B). Nevertheless, the cysteine disulfide (paired with another cysteine containing peptide, P2) was clearly found (P2-P3). In addition, as with the glutathione assignment, the limited backbone cleavage in CID-MS2 was also used for linkage determination (see Figure S2 in the Supplementary Material). Furthermore, identical fragment ions as in Figure 2A (e.g. P1, P2, and P1-P2), which were independent of the glutathione or cysteine linkage, were observed in the spectrum. In summary, the complementary spectra of these two peptide molecules allowed the assignment of the two adjacent cysteines in the peptide backbone and differentiated the unpaired cysteine associated with either glutathione or cysteine. After characterization, the same m/z values of the two species (precursors), measured by FTICR MS, were extracted for comparison. The ratio of glutathione to cysteine on the unpaired cysteine was estimated to be 60/40, assumed that both species had similar response factors, given that the identical long peptide backbone.
It should be noted that the unpaired cysteine with a free sulfhydryl group is reactive, and the pairing with glutathione or cysteine could be protective. Under oxidative or ischemic (hypoxia) stress, the redox of glutathione or cysteine could possibly trigger the pairing molecule to become free (sulfhydryl group) for protein activation, as found for other proteins in biological systems.25
Identification Disulfide Linkages of Peptides #4 and #7 with N-linked Glycosylation
There are three disulfide-linked peptides which each contain one N-linked glycosylation, peptides # 3, 4 and 7, see Figure 1. In principle, PNGase F can be used to remove the N-linked glycosylation or the non-glycosylated counterpart (if partial glycosylation exists) can be directly analyzed to determine the disulfide linkages. However, without reduction, the deglycosylation step by PNGase F can often be inefficient, and two of the three sites in rt-PA are known to be fully glycosylated. Nevertheless, assuming the mass (including glycosylation) of the peptide is not too large (e.g. <10 kDa), our initial strategy was to try to analyze the disulfide-linked peptides with the glycosylation intact.
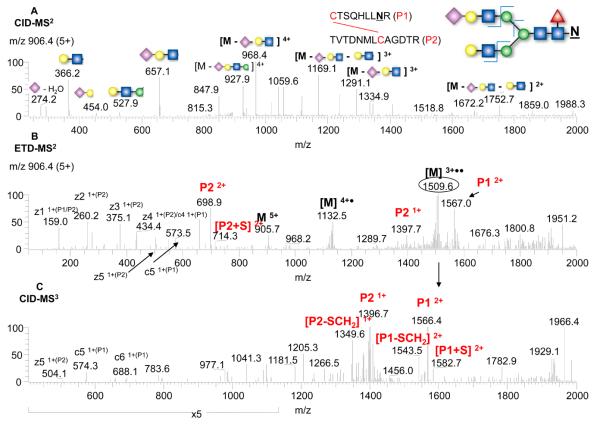
Since ETD fragmentation does not normally produce a glycan fragmentation pattern, the CID-MS2 fragmentation for glycan structure determination for the glycopeptide can be included in the analysis, as shown in Figure 3 for peptide #7. The precursor ion at 906.4 (5+) was fragmented by CID-MS2 (top panel), ETD-MS2 (middle panel), and by CID-MS3 (bottom panel). From the glycosidic bond cleavages in the CID-MS2 along with the precursor mass, a bi-antennary glycan with a terminal sialic acid was assigned, in agreement with our previous results.24 In the ETD-MS2 spectrum, the favorable disulfide bond cleavages were readily seen (i.e. P1 and P2 ions), along with charge-reduced species and peptide backbone fragmented ions (c and z). One of the charge-reduced species ([M+3H]3+••, m/z 1509.6) was isolated and automatically fragmented in the MS3 step, which further confirmed the disulfide linkage assignments from the observation of P1 and P2 ions (bottom panel). Similarly, for the other N-linked glycopeptide which also contained only one disulfide bond linking the two peptides (peptide # 4 in Figure 1), the disulfide assignment was straightforward (see Figures S3A and S3B in the Supplementary Material). In both cases, the disulfide bonds were preferably cleaved by ETD even though the peptides contained different types of glycosyation, i.e. the complex-type (Figure 3) or the high-mannose-type (Figure S3).
Figure 3.
Analysis of the disulfide-linked tryptic peptide with an N-linked glycan by CID-MS2 (A), ETD-MS2 (B), and CID-MS3 (C), with the glycopeptide structure and annotation of the fragment ions indicated in the insert.
In the above analysis, the glycan structure determinations took advantage of the fact that the glycans on rt-PA have been previously studied.26 In the case of unknown glycan structures, the use of PNGase F could simplify the determination of the disulfide assignment, even if the degylcosylation step were not efficient (see Figure S4 in the Supplementary Material). It is interesting to note that we recently showed that glycan removal by PNGase F can be facilitated using pressure cycling technology27, and this approach could potentially aid deglycosylation to simplify disulfide linkage assignments of glycopeptides.
Identification of Disulfide Linkages of Peptide #3 – A Large Glycopeptide
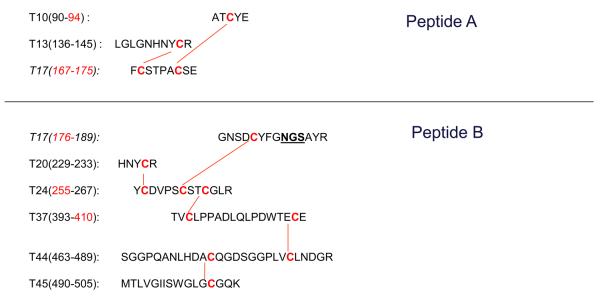
We next examined the remaining disulfide peptide, a glycopeptide with seven disulfides (peptide #3 in Figure 1). This peptide was difficult to analyze because of the large mass, even ignoring glycosylation (~14 kDa). The inefficient fragmentation and wide distribution of charge states for the fragment ions created complexity in the analysis. As a result, we again employed an additional Glu-C digestion step to cut the peptide into smaller pieces. However, as was the case for trypsin digestion, Glu-C could not digest the glutamic acid (E) adjacent to the disulfide (Cys 409 with Cys 484 in Figure 1). As a result, only two peptide fragments were generated, as shown in Figure 4, one with 2 disulfides (~2.7 kDa) and the other with 5 disulfides (~9.5 kDa).
Figure 4.
The structure of the disulfide-linked peptides from a multi-digestion of rt-PA (Lys-C plus trypsin plus Glu-C digestion). The breakage of the peptide with 7 disulfides to 2 fragments are shown, one with 2 disulfides labeled as peptide A, and the other with 5 disulfides labeled as peptide B with the glycosylation site (underlined).
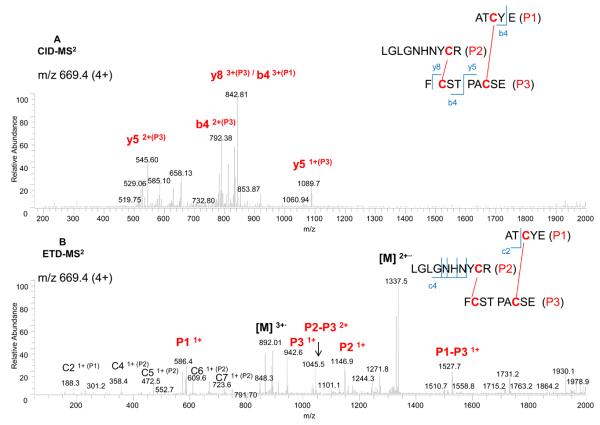
The peptide containing the 2 disulfides (peptide A), m/z of 669.4 (4+), was fragmented by CID-MS2 (top panel) and ETD-MS2 (bottom panel), as shown in Figure 5. As previously, the partially disulfide-dissociated peptides, P1-P3 and P2-P3 in ETD-MS2, could readily be assigned to the linked peptides as P1 with P3 and P2 with P3. The exact linkages (Cys 92 with Cys 173 and Cys 144 with Cys 168) were confirmed by specific fragment ions found in CID-MS2 (i.e. y5 and b4). In this case, the additional confirmation by CID-MS3 was thus not required.
Figure 5.
Analysis of the disulfide-linked peptide A (2 disulfides) from Figure 4 by CID-MS2 (A), and ETD-MS2 (B), with the peptide structure and annotation of the fragment ions indicated in the insert.
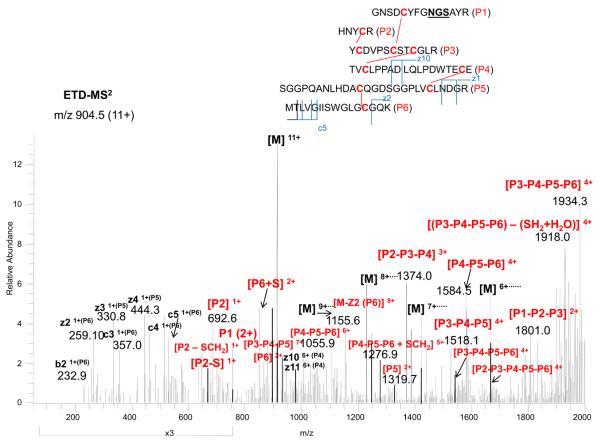
The other peptide with 5 disulfides and an N-glycosylation site (peptide B) was more complex. Since it is known that the glycosylation occupancy at this site is only 50 %,26 the non-glycosylated peptide was chosen for analysis. The peptide precursor m/z, 904.5 (11+), was fragmented by ETD-MS2, as shown in Figure 6. The partially disulfide-dissociated peptides (P1-P2-P3, P2-P3-P4, P3-P4-P5, P4-P5-P6, P3-P4-P5-P6, and P2-P3-P4-P5-P6) could be assigned to the linked peptides, as indicated in the figure. Since many partially linked disulfides were found, the possibilities of other linkages were thus eliminated. It should be noted that we also observed several additional cleavages for this peptide (e.g. further cleaved at D after Glu-C digestion), which could result to the same mass with the addition of water (if the cleaved peptides could still be linked by the disulfides) or additional molecular weight loss (if the cleaved peptides could not be held together by the disulfides). In this case, the assignment of the combination of several miscleavages was difficult initially by matching the precursor masses. The CID fragmentation of these precursor ions were not informative because many fragment ions contained intertwined disulfides still linked together. Nevertheless, the ETD fragmentation provided the clue for assignment since the fragment ions for the non-miscleaved portion were identical, and also these partially cleaved disulfides from the ETD spectrum could be fragmented by CID-MS3for further confirmation. As examples, the CID-MS3 for P2-P3-P4 and P4-P5-P6, as presented in the Supplementary Material (Figures S5 and S6), further confirmed the linkages assigned from the ETD spectrum. Thus, as demonstrated here, the analysis of multiple intertwined disulfides in a peptide requires the combination of multiple enzymes with sequential digestion, coupled with ETD-MS2 and targeted CID-MS3.
Figure 6.
ETD-MS2 spectrum of the disulfide-linked peptide B (5 disulfides) from Figure 4, with the peptide structure and annotation of the fragment ions indicated in the insert.
Analysis of TNK-tPA
After the complete characterization of disulfides in rt-PA, we also examined the disulfide linkages and the status of the free cysteine in the rt-PA mutant (TNK-tPA or Tenecteplase), which is also approved for the same indications as rt-PA (myocardial infarction and acute ischemic stroke). TNK-tPA has been shown to have a longer half life and higher fibrin-binding specificity than rt-PA because of the difference in glycosylation patterns for the two therapeutics.22-23 Although TNK-tPA has amino acid substitutions at 3 sites (see Experimental Section), the amino acid sequences remain identical for all disulfide-linked peptides for both forms of t-PA. Because the masses of the disulfide-linked peptides are identical, the determination of TNK-tPA disulfides was straightforward by comparing the masses to those found for rt-PA. Importantly, we again found that the free sulfhydryl group for TNK-tPA was fully blocked by glutathione and cysteine at the similar 60/40 ratio.
It is important to note that rt-PA and TNK-tPA were obtained from different clones and thus from different cell cultures and down-stream manufacturing conditions. However, neither the glycosylation (caused by the point mutation) nor the manufacturing conditions appear to influence the disulfide formation; rather the amino acid sequences in close proximity to the cysteine residues appear to dictate disulfide formation during protein folding. Thus, TNK-tPA is a well engineered mutant, where it is only mutated sufficiently to change the outer surface glycans while not influencing the folding disulfides, even at the location of the unpaired cysteine (which may influence activation upon oxidative or ischemic stress).
Conclusions
The 17 disulfides of rt-PA were completely assigned, for the first time, by the combination of multiple and sequential enzymatic digestion along with the CID/ETD and CID-MS3. The result is consistent with homology predictions. Additionally, the unassigned (unpaired) cysteine, identified at position 83, was found to be paired with a glutathione or cysteine molecule. As we have noted, the identification of the location and status of the unpaired cysteine should enable the study of the activation or signaling of rt-PA upon stimulation by oxidative or hypoxia stress, such as for patients under ischemic stroke (hypoxia) or myocardio-infarction bleeding (oxygen exposure).
In the production of therapeutic recombinant proteins, the analysis demonstrated in this paper should provide important confirmatory evidence that the protein is folded properly. In continuation of this work, we are at present developing with colleagues a software tool to simplify disulfide assignments and also to guide users for predicting critical cleavages for confirmation.
Supplementary Material
Acknowledgement
The authors acknowledge NIH for support of this work, GM 15847 (BLK) and CA128427 (WSH). The authors further thank Genentech for the gift of rt-PA and TNK-tPA. Contribution Number 963 from the Barnett Institute.
References
- 1.Wu SL, Kim J, Hancock WS, Karger BL. J Proteome Res. 2005;4:1155–67. doi: 10.1021/pr050113n. [DOI] [PubMed] [Google Scholar]
- 2.Wu SL, Kim J, Bandle RW, Liotta L, Petricoin E, Karger BL. Mol Cell Proteomics. 2006;5:1610–27. doi: 10.1074/mcp.M600105-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Zheng X, Wu SL, Hancock WS. Int J Pharm. 2006;322:136–45. doi: 10.1016/j.ijpharm.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 4.Jiang H, Wu SL, Karger BL, Hancock WS. Biotechnol. Prog. 2009;25:207–18. doi: 10.1002/btpr.72. [DOI] [PubMed] [Google Scholar]
- 5.Schellekens H. J Nephrol. 2008;21(4):497–502. [PubMed] [Google Scholar]
- 6.Mamathambika BS, Bardwell JC. Annual Review of Cell and Developmental Biology. 2008;24:211–35. doi: 10.1146/annurev.cellbio.24.110707.175333. [DOI] [PubMed] [Google Scholar]
- 7.Bardwell JC, Beckwith J. Cell. 1993;74(5):769–71. doi: 10.1016/0092-8674(93)90455-y. Review. [DOI] [PubMed] [Google Scholar]
- 8.Giles NM, Giles GI, Jacob C. 2003;300:1–4. doi: 10.1016/s0006-291x(02)02770-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Boyle PC, Lu BY, Chang JY, Wriggers W. Biochemistry. 2006;45:15269–78. doi: 10.1021/bi0615083. [DOI] [PubMed] [Google Scholar]
- 10.Tu SC, Ho-Schleyer SC, Travers KJ, Weissman JS. Science. 2000;290:1571–74. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 11.Forman HJ, Fukuto JM, Torres M. Am J Physiol Cell Physiol. 2004;287:C246–C256. doi: 10.1152/ajpcell.00516.2003. Review. [DOI] [PubMed] [Google Scholar]
- 12.Haniu M, Acklin C, Kenney WC, Rohde MF. Int J Pept Protein Res. 1994;43(1):81–6. doi: 10.1111/j.1399-3011.1994.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, Cockrill SL. Anal. Chem. 2009;81:7314–20. doi: 10.1021/ac901161e. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Kaltashov IA. Anal Chem. 2006;78:4820–9. doi: 10.1021/ac060132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo JA, Edmonds CG, Udseth HR, Smith RD. Anal Chem. 1990;62:693–8. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 16.Bean MF, Carr SA. Anal Biochem. 1992;201:216–26. doi: 10.1016/0003-2697(92)90331-z. [DOI] [PubMed] [Google Scholar]
- 17.Riener CK, Kada G, Gruber HJ. Anal Bioanal Chem. 2002;373:266–76. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 18.Chrisman PA, Pitteri SJ, Hogan JM, McLuckey SA. J Am Soc Mass Spectrom. 2005;16:1020–30. doi: 10.1016/j.jasms.2005.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Anal Chem. 2000;72:563–73. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 20.Anusiewicz I, Berdys-Kochanska J, Simons J. J Phys Chem A. 2005;109:5801–13. doi: 10.1021/jp050218d. [DOI] [PubMed] [Google Scholar]
- 21.Wu SL, Jiang H, Lu Q, Dai S, Hancock WS, Karger BL. Anal Chem. 2009;81:112–22. doi: 10.1021/ac801560k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benedict CR, Refino CJ, Keyt BA, Pakala R, Paoni NF, Thomas GR, Bennett WF. Circulation. 1995;92(10):3032–40. doi: 10.1161/01.cir.92.10.3032. [DOI] [PubMed] [Google Scholar]
- 23.Keyt BA, Paoni NF, Refino CJ, Berleau L, Nguyen H, Chow A. Proc. Natl. Acad. Sci. 1994;91:3670–3674. doi: 10.1073/pnas.91.9.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu SL, Hühmer AF, Hao Z, Karger BL. J Proteome Res. 2007;6:4230–44. doi: 10.1021/pr070313u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryser HJ, Levy EM, Mandel R, DiSciullo GJ. Proc. Natl. Acad. Sci. 1994;91:4559–63. doi: 10.1073/pnas.91.10.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spellman MW, Basa LJ, Leonard CK, Chakel JA, O'Connor JV, Wilson S, van Halbeek H. J Biol Chem. 1989;264(24):14100–11. [PubMed] [Google Scholar]
- 27.Szabo Z, Guttman A, Karger BL. Anal Chem. 2010;82:2588–93. doi: 10.1021/ac100098e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.