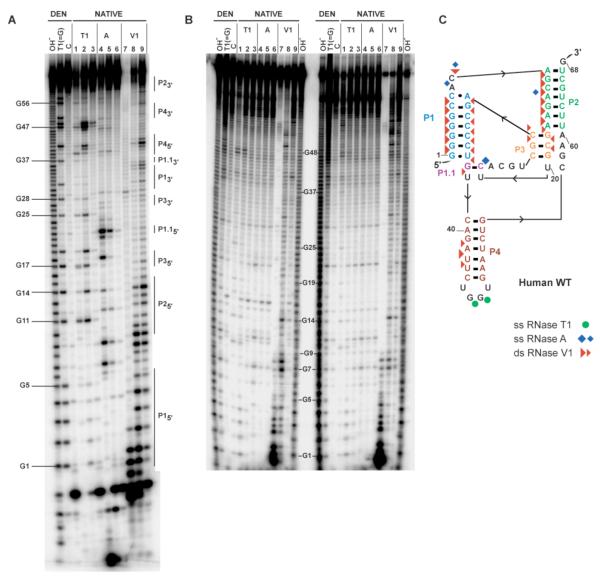
FIGURE 3.
Enzymatic structure mapping of human WT CPEB3 ribozyme. (A) Structure mapping of self-cleaved 1/68 WT ribozyme. Denaturing 12% polyacrylamide gel is shown. 5′- end-labeled RNA was treated with RNases as described in Materials and Methods. The first three lanes are denaturing conditions (DEN), and the others are native conditions (NATIVE). Labels are as follows; OH−, limited alkaline digest; T1, A, and V1 limited digests with RNases T1 (G-specific), A (pyrimidine-specific), and V1 (double strand-specific), respectively; C, control sample without enzyme. Sets of three decreasing concentrations of each ribonuclease are provided (see Materials and Methods for details), and those with single hit kinetics were used for structure assignments, which is the third lane in each set. Numbering refers to selected bands produced by limited digestion with RNase T1. For the self-cleaved ribozyme, positions of the pairing elements are delineated by vertical lines and labeled on the right-hand side. Note that RNase V1 cleavage products greater than ~10 nt migrate approximately 1 nt slower than other cleavages owing to absence of a 2′,3′-cyclic phosphate. Fragments shorter than 10 nt generated by RNase V1 cleavages have anomalous migrations relative to RNase T1 and hydrolysis lanes (41). There are additional bands at the very bottom of the gel (not shown) that did not align with the T1 ladder and likely represent 2′ and/or 3′ phosphate monoesters. (B) Structure mapping of precursor–8/68 WT ribozyme. The gel is labeled as described above for the cleaved ribozyme. Selected bands produced by limited digestion with RNase T1 are numbered. Lanes to the left of the numbering are for precursor RNA band R1, and those to the right are for band R2 (see Figure 2). Both of these bands had been purified from a native gel as described in Materials and Methods. (C) Single and double stranded cleavages for the self-cleaved form in panel A mapped to a secondary structure. This structure is largely consistent with structure mapping of the self-cleaved form of the ribozyme and with free energy minimizations using ILM (29, 30), as well as the structure proposed by Salehi-Ashtiani et al (14). Positions of cleavage by RNases T1, A, and V1 are shown using green circles, blue diamonds, and red triangles respectively. Symbol size is relative to cleavage intensity.