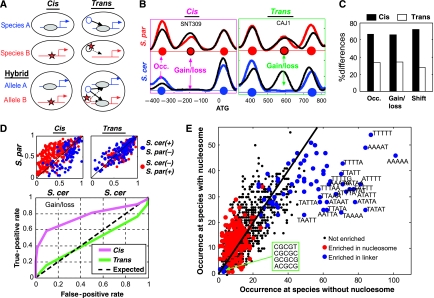
Figure 2.
Cis- and trans-dependent changes in nucleosome positioning. (A) An inter-species difference in nucleosome positioning that is generated in cis will be preserved within the hybrid. In contrast, an inter-species difference in nucleosome positioning that is generated in trans will not be preserved in the hybrid as the two hybrid alleles are at the same nucleus and thus affected by the same proteins in trans. (B) Examples of changes classified as cis and trans, showing nucleosome scores (smoothed nucleosome occupancy) of S. cerevisiae (blue), S. paradoxus (red) and the corresponding hybrid alleles (black). Predicted center locations of nucleosomes are marked with circles. (C) Estimation of the percentage of cis and trans differences (see Materials and methods). (D) Differences in cis (but not trans) are predicted from sequence divergence. Nucleosome occupancy was predicted in the two species with a sequence-based model (Field et al, 2008). Upper panel: the predicted nucleosome occupancy of S. cerevisiae (x axis) and S. paradoxus (y axis) at positions of nucleosome gain/loss; blue and red dots refer to nucleosomes that are present only in S. cerevisiae and S. paradoxus, respectively. Lower panel: predictive power of the sequence model. For each possible threshold, we examined the frequency of nucleosome gains/losses with difference in predicted occupancy that pass the threshold (true positives) and compared it with the frequency of randomly selected positions (false positives) with difference in predicted occupancy that pass this threshold. (E) The number of times each 5-mer is found at positions of nucleosome gains/losses at the species without (x axis) or with nucleosome (y axis). Sequences are indicated for the most significantly biased 5-mers. Colors indicate whether a 5-mer is generally associated (in S. cerevisiae) with linker regions (blue) or nucleosomes (red).