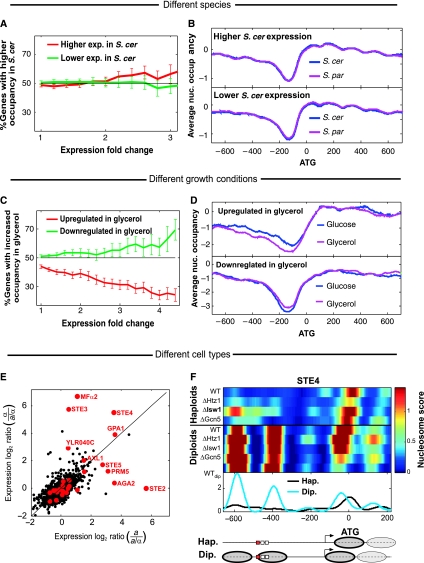
Figure 5.
Divergence of nucleosome positioning is not correlated with divergence of gene expression. (A) Percentage of genes with higher promoter nucleosome occupancy in S. cerevisiae (compared with S. paradoxus) among all the genes with significantly higher or lower expression in S. cerevisiae, as a function of the threshold for differential expression. Note that the slight (not statistically significant) trend toward higher nucleosome occupancy of genes with higher expression is opposite to the expected trend. (B) Average nucleosome occupancy for all genes with 1.5-fold higher expression in S. cerevisiae (top) or 1.5-fold lower expression in S. cerevisiae (bottom), compared with S. paradoxus. (C, D) Significant correlation between changes in nucleosome positioning and changes in gene expression for comparison of S. cerevisiae grown on glucose and glycerol. Figures are as described in (A, B) but for comparison of the same species in two conditions. (E) Differential expression between haploids and diploids for S. cerevisiae (a versus a/α) and S. paradoxus (α versus a/α). Genes with similar nucleosome positioning in haploids and diploids are shown in black and genes with the most significant differences in nucleosome positioning are shown in red (differences were averaged over the different species and strains, see Materials and methods for definition); 10 of the 28 genes (36%) with differential haploid–diploid positioning have at least fourfold higher expression in one of the haploids than the diploids and their gene names are indicated. In contrast, only 2% of the genes with similar positioning display such haploid-specific expression (P<10−16, binomial test). (F) Patterns of nucleosome positioning in haploids and diploids, for the haploid-specific gene STE4. Data are shown for four S. paradoxus haploid α strains, and for four hybrid and one S. paradoxus (WTdip) diploid a/α strains. Curves represent the average nucleosome scores of haploids (black) and diploids (cyan). Predicted nucleosome positions, TSS and binding sites (red for STE12 sites and white for others) are indicated below. Note that deletion of ISW1 decreases the difference between haploids and diploids, implicating ISW1 in this differential nucleosome positioning (see also Mfα2 and YLR040C at Supplementary Figure S10).