Abstract
PIK3CA mutations are reported to be present in approximately 25% of breast cancer (BC), particularly the estrogen receptor–positive (ER+) and HER2-overexpressing (HER2+) subtypes, making them one of the most common genetic aberrations in BC. In experimental models, these mutations have been shown to activate AKT and induce oncogenic transformation, and hence these lesions have been hypothesized to render tumors highly sensitive to therapeutic PI3K/mTOR inhibition. By analyzing gene expression and protein data from nearly 1,800 human BCs, we report that a PIK3CA mutation–associated gene signature (PIK3CA-GS) derived from exon 20 (kinase domain) mutations was able to predict PIK3CA mutation status in two independent datasets, strongly suggesting a characteristic set of gene expression–induced changes. However, in ER+/HER2− BC despite pathway activation, PIK3CA mutations were associated with a phenotype of relatively low mTORC1 signaling and a good prognosis with tamoxifen monotherapy. The relationship between clinical outcome and the PIK3CA-GS was also assessed. Although the PIK3CA-GS was not associated with prognosis in ER− and HER2+ BC, it could identify better clinical outcomes in ER+/HER2− disease. In ER+ BC cell lines, PIK3CA mutations were also associated with sensitivity to tamoxifen. These findings could have important implications for the treatment of PIK3CA-mutant BCs and the development of PI3K/mTOR inhibitors.
Keywords: gene expression profiling, PI3 kinase
Deregulated PI3K-AKT signaling has been implicated in many aspects of carcinogenesis. Many genomic alterations have been found to activate this pathway, which can contribute to tumor progression, metastases and resistance to treatment (1). In particular, somatic mutations of the PIK3CA gene, encoding the p110α catalytic subunit of PI3K, have been shown to activate AKT and induce oncogenic transformation in vitro and in vivo (2–4). Mutations of the PIK3CA gene have been found in 18% to 40% of human breast cancers (BCs) (5–7), making them one of the most common genetic aberrations in human BC; however, their precise downstream effects have not been clearly elucidated (1, 3).
In BC, the relationship between PIK3CA mutations with clinical outcome remains unclear (8). This is most likely a result of the mostly small and heterogeneous patient populations studied in previous reports (7, 9,10,11,12–13), as the clinical consequences of PIK3CA mutations may depend on a patient's BC molecular subtype [e.g., HER2+ vs. estrogen receptor (ER)–positive/HER2−], the treatments received, and the presence of other oncogenic aberrations. Despite the many uncertainties surrounding the complex biology of PI3K signaling, a large number of PI3K inhibitors—specifically targeted to tumors possessing PI3K genetic aberrations such as PI3KCA mutations—are currently under development as potential anticancer compounds.
In previous studies of gene expression profiling in human BC, two distinct molecular subgroups of ER+/HER2− BC could be defined. These subgroups, initially referred to as Luminal- A and -B (14), were associated with significantly different prognoses: good and poor, respectively (15–18). Because expression of proliferation genes seems to define the clinical phenotype—high proliferation rates equates with poor outcome, in particular with tamoxifen monotherapy (16)—we hypothesized that PIK3CA mutations could be responsible for the oncogenic deregulation in the poor prognostic group and consequently these patients would be most suitable for PI3K/mammalian target of rapamycin (mTOR) pathway inhibition.
Therefore, the specific objective in this study was to better characterize the clinical relevance and molecular changes associated with a PIK3CA mutation in ER+/HER2− BC. To do this, we defined a gene-expression signature driven by PIK3CA exon 20 [kinase domain (KD)] mutations and show here that, surprisingly, in human ER+/HER2− BC, PIK3CA mutations were associated with relatively low mTORC1 functional output and a good clinical outcome with adjuvant tamoxifen monotherapy.
Results
PIK3CA Mutant ER+ BCs Are Associated with a Distinct Gene Expression Signature.
Mutational analysis of the PIK3CA gene was performed in 173 primary ER+/HER2− BCs (TAM dataset; Table S1). A total of 46 mutations were found (26%), the majority located on the KD, with 91% of these being H1047R substitutions (Table S2). PIK3CA mutations detected by sequencing were not significantly correlated with any other important clinicopathologic features (Table S3). To determine whether PIK3CA mutations induced characteristic transcriptional changes, we compared gene expression profiles between PIK3CA mutant (mt) and WT samples using a two-sample t test. Only PIK3CA KD mt tumors were used in this analysis to exclude the possibility that mutations in different exons might produce different biological phenotypes. We found 278 probe sets with a fold change of >1.3 and a P value <0.05 (Dataset S1). The probability of obtaining this gene signature by chance was 0.02 after 1,000 random permutations of the class labels.
We hypothesized that a gene-expression signature associated with PIK3CA mutations would help us to better understand their effect on the pathway and their relationship to clinical outcome. To develop a predictive gene signature score, we took these probe sets and computed the sum of the average of the logarithmic gene expression—hereafter referred to as PIK3CA-GS—for each BC sample. We then assessed the ability of the PIK3CA-GS to classify BCs that were PIK3CA mt or WT by sequencing. In the TAM dataset on which the signature was developed, leave-one-out cross-validation of the procedure was performed and good classification performance was observed (area under the curve [AUC] of the receiver operating curve, 0.74; P < 0.0001).
To independently assess the ability of the PIK3CA-GS to determine PIK3CA mutation status, we evaluated two separate cohorts of patients with BC—105 and 129, respectively—with known PIK3CA mutation status (19, 20). BCs that were known to be HER2-amplified were excluded in these analyses as these BCs would already exhibit PI3K pathway activation. We observed similar predictive ability in both datasets containing both ER+ and ER− samples, with both PIK3CA HD and KD mutations [Saal et al. (19) dataset, AUC, 0.7, P = 0.003; M. D. Anderson Cancer Center (MDACC) dataset (20), AUC, 0.71, P = 0.002; Fig. S1]. Although we did not develop the PIK3CA-GS to replace sequencing for detection of PIK3CA mutant status, these results confirmed that, in human BCs, PIK3CA mt and WT tumors are associated with distinct gene expression profiles. We also noticed that regardless of the genes used, as many as 20% of PIK3CA WT tumors were misclassified as mt during the leave-one-out process. This suggests that other PI3K pathway aberrations may produce a phenotype similar to that of a PIK3CA mutation. Also of interest were the five AKT1 (3.8%) mutations (all E17K) also detected in this dataset (21). The PIK3CA-GS predicted AKT1 mutations (AUC, 0.77; P = 0.04), indicating a similar pattern of gene expression. As PIK3CA and AKT1 mutations were mutually exclusive, this would exclude any possibility of over-fitting the data.
Correlation of PIK3CA Mutations and the PIK3CA-GS with Clinical Outcome in Human BC According to Molecular Subtype.
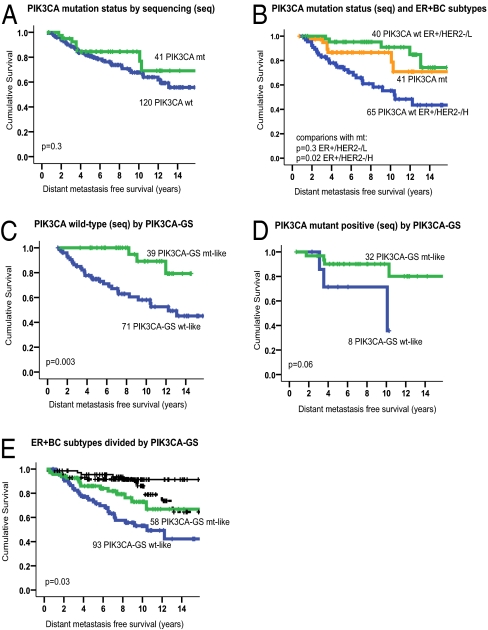
Patients with known PIK3CA mutations by sequencing did not have a significantly worse prognosis compared with those without mutations (TAM dataset; Fig. 1A). Also of note, all AKT1 mt ER+ BC patients remained relapse-free after 7 y of follow-up. PIK3CA HD and KD mutations were examined combined and separately and results were similar (Table S4). However, compared with the prognostic subgroups known to exist in ER+/HER2− BC defined according to their expression of proliferation genes (16), PIK3CA mutants had a similar survival compared with the good-prognosis, low-proliferative (ER+/HER2−/L) subgroup and a better outcome compared with the poor-prognosis, highly proliferative (ER+/HER2−/H) subtype (P = 0.03; Fig. 1B). It is important to note that all patients in this analysis had received the same adjuvant treatment, thereby eliminating interactions between different treatments as potential confounding factors on patient outcome.
Fig. 1.
The PIK3CA-GS has better prognostic significance than mutation status alone. Kaplan-Meier curves according to PIK3CA mutation status by sequencing or by the PIK3CA-GS in (A) all patients in whom mutation status was assessed, (B) PIK3CA mt compared with prognostic subgroups of ER+ BC, (C) patients classified as PIK3CA WT by sequencing, (D) patients classified as PIK3CA mt by sequencing, and (E) ER+ BC classified as good prognosis (black: ER+/HER2−/L) and poor prognosis (green/blue: ER+/HER2−/H) molecular subtypes and prognosis according to PIK3CA-GS expression.
Hypothesizing that the PIK3CA-GS would be able to provide useful information about the activity of the PI3K/AKT pathway, even in patients with PIK3CA WT BC, the PIK3CA-GS was evaluated by using BC samples with unknown PIK3CA mutation status. Note that clinical outcome was not used to develop the signature, and hence all the survival analyses were unbiased in their estimate of its prognostic performance. The abilities of the PIK3CA-GS were also examined separately in each BC molecular subtype (22).
As shown, the PIK3CA-GS was not associated with clinical outcome in the HER2+ and ER−/HER2− (i.e,. “triple negative”) subgroups of patients with BC who had received no systemic treatment [untreated (UNT) dataset], thus providing a “clean” group for prognostic assessment (Table 1). However, in the ER+/HER2− group (n = 717), those tumors classified as “mt-like” or with a gene expression profile similar to that of a PIK3CA KD mutation had a significantly better outcome than those classified as “WT-like.” As all patients with ER+ BC today receive adjuvant hormonal therapy, the prognostic ability of the PIK3CA-GS in tamoxifen-treated ER+ BC is shown in Table 2. Of note, the PIK3CA-GS had prognostic ability in both ER+ BCs known to be PIK3CA WT and mt by sequencing. Of high clinical interest was the PIK3CA-GS's association with a better outcome in the ER+/HER2−/H subgroup, the prognosis of which with tamoxifen treatment is normally poor.
Table 1.
Cox univariate analysis in the three BC molecular subtypes in patients who received no systemic treatment
| Univariate Cox* analysis | |||
| BC subtype | No. of samples | HR (95% CI) | P Value |
| Triple negative | 280 | 0.7 (0.4–1.1) | 0.3 |
| HER2-overexpressing | 192 | 1.3 (0.6–2.7) | 0.4 |
| ER+/HER2− | 717 | 0.7 (0.4–0.8) | 0.00002 |
All BC subtypes are defined using gene expression. HR, hazard ratio; seq, mutation status by sequencing.
*PIK3CA-GS treated as a continuous variable (cut-off independent).
Table 2.
Cox univariate HRs from patients with ER+ BC who received tamoxifen monotherapy
| Univariate Cox* analysis | |||
| ER+ BC treated with tamoxifen monotherapy | No. of samples | HR (95% CI) | P Value |
| All ER+ BC | 323 | 0.4 (0.2–0.6) | 0.00001 |
| PIK3CA WT (seq) | 110 | 0.3 (0.2–0.6) | 0.001 |
| PIK3CA mutant (seq) | 40 | 0.1 (0.01–0.7) | 0.02 |
| ER+/HER2−/low proliferative subtype | 161 | 0.6 (0.2–1.9) | 0.4 |
| ER+/HER2−/high proliferative subtype | 162 | 0.5 (0.3–0.9) | 0.03 |
All BC subtypes are defined using gene expression. HR, hazard ratio; seq, mutation status by sequencing.
*PIK3CA-GS treated as a continuous variable (cut-off independent).
The Kaplan-Meier curves in Fig. 1 C and D illustrate that the functional read-out of the PI3K pathway provided by the PIK3CA-GS can provide outcome information in ER+ BC treated with tamoxifen in both PIK3CA sequenced WT and mt samples, respectively. Two prognostic groups were generated by ranking patients according to the PIK3CA-GS and dividing at the median level. Fig. 1E demonstrates the ability of the PIK3CA-GS to divide the each ER+ BC subtype. Multivariate analysis confirmed that the PI3KCA-GS could provide independent prognostic information in the tamoxifen-treated patients (Table 3) and PIK3CA sequenced WT group (P = 0.06). Overall, our data suggest that the PIK3CA-GS has prognostic abilities, particularly for the ER+/HER2− BC, and that the PIK3CA-GS was a stronger prognostic indicator than mutation status alone.
Table 3.
Multivariate analysis in patients with ER+ BC who had received adjuvant tamoxifen (TAM dataset, n = 302)
| Factor | HR (95% CI) | P Value |
| Age (binary) | 1.9 (0.4–8.1) | 0.4 |
| Nodal status | 1.6 (0.8–3.1) | 0.2 |
| Tumor size (binary) | 1.9 (1.1–3.3) | 0.03 |
| Histologic grade | 2.4 (0.9–6.2) | 0.07 |
| PIK3CA-GS | 0.5 (0.3–0.8) | 0.01 |
Patients were included from the TAM dataset who had all data available for analysis. All BC subtypes are defined using gene expression; HR, hazard ratio. In multivariate analysis, factors were treated as follows: age ≤50 y vs. >50 y; nodal status: positive vs. negative; tumor size: ≤2 cm vs. >2 cm; histologic grade 3 vs. 1 and 2. Endpoint is distant metastases.
*PIK3CA-GS treated as a continuous variable (cut-off independent).
PIK3CA Mutations in ER+/HER2− BC Are Associated with a Gene Signature of PI3K/AKT Pathway Activation but Relatively Low mTORC1 Signaling Output.
We next sought to better understand the biological signal associated with the gene expression pattern of PIK3CA mutations in human ER+/HER2− BC, given that it was able to predict mutation status in independent datasets, yet was correlated with a relatively good prognosis in 1,071 ER+ BC samples. These results seemed contrary to studies in experimental systems that have indicated (i) that activating mutations in PIK3CA results in increased neoplastic transformation through the PI3K pathway, and (ii) that increased levels of phosphorylated AKT have been associated with poor clinical outcome in BC (2, 4, 23).
Considering recent reports that PIK3CA mutations can induce cellular senescence, we looked for evidence of this using senescence-associated β-Gal (SA-β-Gal) staining (24, 25). For the subset tested, we found no evidence of increased senescence in PIK3CA mt or PIK3CA-GS classified mt-like samples compared with WT. Next, we used Ingenuity Pathways Analysis (IPA) to compare the mt and WT samples for interactions of the differentially expressed genes. In the mt samples we noted significantly increased expression of PIK3R1 and PIK3R3 and decreased expression of IRS2, but, surprisingly, also of RPS6K, RHEB, EIF4E, many ribosomal and eurokarytic translation protein genes, and downstream cMYC (Fig. S2). These patterns were also apparent in the mt-like (PIK3CA-GS classified) samples.
In a further comparison of PIK3CA mt and WT samples, we used GSEA (26) and curated gene sets obtained from Biocarta and the literature of PI3K/AKT/mTOR activation. Only an AKT-regulated gene set derived from Biocarta was significantly enriched in the PIK3CA mt samples (P = 0.02; false discovery rate, 0.1). Using GSEA in patient groups stratified by the PIK3CA-GS, we found similar results: the AKT-regulated genes were also enriched in the mt-like group (P = 0.01). However, the PIK3CA-GS was negatively correlated with gene signatures of proliferation (R = −0.4; P < 1−6) (27), AKT activation (R = −0.1; P = 0.03) (28), and PTEN loss (R = −0.4; P < 1−6) (19) but positively correlated with ESR1 expression (R = 0.3; P < 1−6).
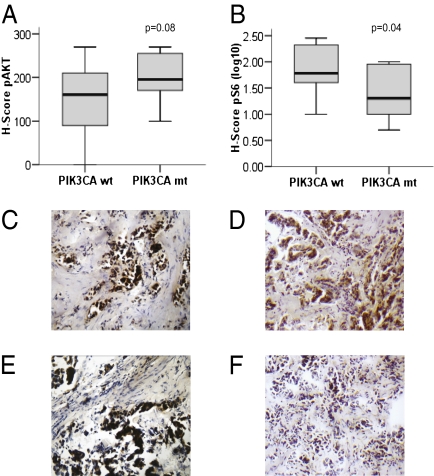
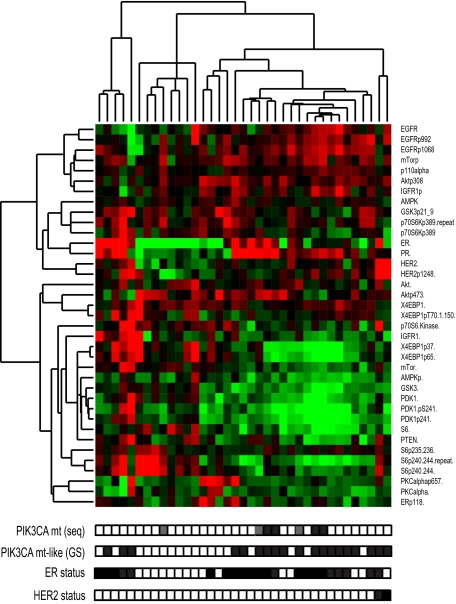
Given the importance of posttranslational regulation of the PI3K/AKT/mTOR pathway, it was critical to confirm these downstream findings at the protein level. In the TAM dataset, we performed immunohistochemistry (IHC) with antibodies to phosphorylated AKT and S6. In the PIK3CA WT compared with mt samples, respectively, we found pAKT expression was nonsignificantly up-regulated (P = 0.08) whereas pS6 expression was significantly down-regulated (P = 0.04; Fig. 2). Importantly, these findings were also observed in the independent dataset from MDACC, in which protein expression was evaluated by using reverse-phase protein arrays (Fig. 3 and Dataset S2). As seen, when unsupervised hierarchical clustering was performed, the mt and mt-like samples cluster together. The protein data were consistent with gene expression data showing decreased expression of the downstream proteins mTOR, S6K, and S6 associated with less phosphorylation of 4EBP1 and S6, further supporting the concept that the PIK3CA-GS was associated with relatively reduced downstream signaling in the mt and mt-like groups.
Fig. 2.
Box-plots comparing expression of (A) p-AKT and (B) p-S6 levels in PIK3CA mt and WT samples from the TAM dataset. Examples of PIK3CA mt and WT samples (C and E, respectively): pAKT was present in both the nuclei and cytoplasm, and pS6 staining was observed mainly in the cytoplasm from positive tissue specimens (D and F, magnification 400×, high-power field).
Fig. 3.
PIK3CA mt samples and mt-like samples have show decreased expression of downstream PI3K/AKT/mTOR proteins. Heat map represents unsupervised hierarchical clustering of BCs and corresponding protein data from the MDACC cohort. Patients are represented horizontally: PIK3CA mt status by sequencing (seq), according to gene signature (GS), ER and HER2 (FISH) status are indicated by filled (positive) and white (negative) boxes and Exon 9 and 20 mutations are gray and black boxes, respectively; Clustering was performed by using Pearson correlation metric and centroid linkage. Proteins and phosphorylated proteins are indicated vertically. Red represents over-expression, green represents underexpression.
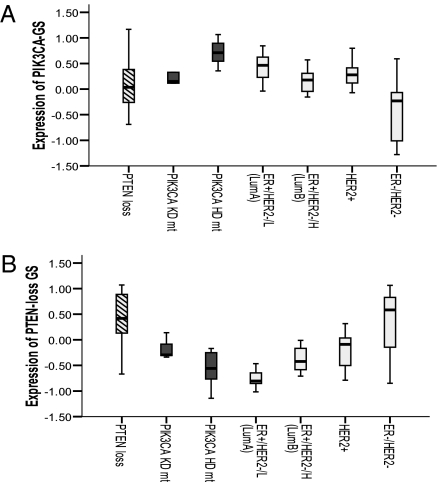
Finally, in light of recent data suggesting that not all PI3K pathway alterations are comparable (8, 29), we asked whether we were seeing “relatively low” mTORC1 signaling as a result of PIK3CA mutations being “weak” activators of the PI3K pathway. Fig. 4 demonstrates significant differences in the expression of the PIK3CA-GS and PTEN-loss gene signatures—both of which summarize PI3K pathway activation but are driven by different aberrations—in BCs that divided according to PTEN deficiency (on IHC), PIK3CA-mutated, HER2-amplified, and molecular subtypes. Notable is the contrast between PTEN-loss and PIK3CA-mutated BCs. Notably, PIK3CA mts only have higher pathway activation compared with the Luminal A subtype (Fig. 4B). Together these data suggest that, in ER+/HER2− BC with PIK3CA mutations, pathway activation surprisingly does not result in greatly elevated downstream signaling and their functional output differs substantially compared with that of PTEN loss.
Fig. 4.
Box plots showing relative expression of the PIK3CA mt (A) and PTEN-loss (B) gene signatures according to molecular subtype and PI3K pathway aberration from the Saal et al. (19) dataset. PIK3CA-sequenced mutants (HD+KD) have less expression of the PTEN-loss gene signature compared with BCs that are PTEN-deficient by IHC (P = 0.001) or HER2-amplified (P = 0.01), but still higher expression compared with the ER+/HER2−/L (Luminal-A) tumors (P = 0.002).
Correlation of PIK3CA Mutation Status and PIK3CA-GS in ER+ BC Cell Lines to Tamoxifen and Rapamycin Response.
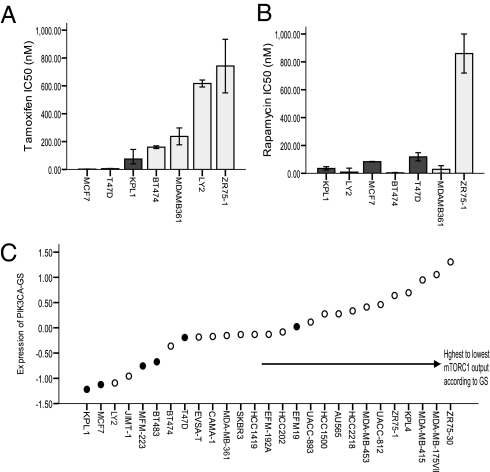
We next evaluated sensitivity to tamoxifen in a panel of seven ER+ cell lines with known PIK3CA mutation status (Fig. 5A). As seen, PIK3CA mt cell lines were associated with sensitivity to tamoxifen, corroborating the clinical outcome data. In contrast in the same cell lines, PIK3CA mutation status did not correlate with rapamycin sensitivity (Fig. 5B). However, when the cell lines are ranked according to increasing PIK3CA-GS values (Fig. 5C), the cell line with the highest value of the PIK3CA-GS—the lowest mTORC1 output—was the most rapamycin resistant (ZR751). Of note, the PIK3CA-GS was able to discriminate PIK3CA mutation in status in cell lines using publicly available data (30). However, in cell lines the signature was significantly predictive but in the reverse direction compared with human PIK3CA mt ER+ BCs (AUC = 0.9; P = 0.008; Fig. S1C), i.e., PIK3CA mt cell lines had the lowest values of the PIK3CA-GS correlating with high mTORC1 output (Fig. 5C). This supports the notion that the strength of pathway activation may differ in vitro compared with human PIK3CA mt BC. We also queried the “Connectivity Map” (31) and found that, similar to our results described earlier, the PIK3CA-GS was positively correlated, i.e., had gene expression profiles similar to that induced by rapamycin (0.45) in MCF7 cell lines. Overall, these results reinforce that (i) PIK3CA mutations may impart a better response to tamoxifen, (ii) the PIK3CA-GS is associated with mutation status both in human BCs and in cell lines, and (iii) PIK3CA-GS could be helpful as a clinical biomarker of the functional PI3K/mTOR output.
Fig. 5.
In a panel of ER+ BC cell lines, IC50 was assessed to rapamycin and tamoxifen according to PIK3CA status. PIK3CA mutations (MCF7, T47D, KPL1) predict sensitivity to tamoxifen (A), but not rapamycin (B). Cell lines are ranked according to increasing PIK3CA-GS (C) The PIK3CA-GS could identify ER+ PIK3CA mt lines, but in the reverse direction to human BC (AUC, 0.9; P = 0.008; Fig. S1C), i.e., cell lines are associated with relatively high mTORC1 output. The rapamcyin-resistant ZR751 line was associated with the highest PIK3CA-GS value; black bars/circles represent PIK3CA mt cell lines in A, B, and C; error bars indicate 95% CI; each bar represents the mean IC50 from triplicate plates.
Discussion
In the present study, we sought to determine whether PIK3CA mutations could be the oncogenic driver responsible for the poor prognosis of the highly proliferative ER+/HER2− BC phenotype. To our surprise, we found that PIK3CA mutations were not associated with a poor clinical outcome despite their known tumorigenic effects through activation of the PI3K pathway. We observed that, whereas PIK3CA mutations were associated with a distinct gene expression signature of PI3K pathway activation—confirmed in two independent datasets of human BC—in contrast, the PIK3CA-GS could identify ER+/HER2− BC patients, both untreated and tamoxifen-treated, with better outcomes. Moreover this ability was independent of their mutation status. We also report that PIK3CA-GS was not prognostic in ER− or HER2+ BC subtypes, suggesting that the molecular background of BC is an important determinant of the functional output of a PIK3CA mt BC. Finally we show that in ER+/HER2−/PIK3CA mt BCs, despite apparent PI3K/AKT pathway activation, downstream mTORC1 signaling was not greatly elevated at the transcriptional and biochemical levels. These patterns were observed from the gene expression data as well as corresponding protein data from multiple independent datasets of human BC.
Recently, the PIK3CA mutation has been described as a “good activating mutation” (32) given increased reporting of its association with favorable prognostic features such as ER positivity and a relatively good clinical outcome with hormonal therapy (8, 9, 12, 13, 33). Our data support this but suggest that the effect is restricted to ER+/HER2− BC. It was previously reported that PIK3CA mt cell lines were sensitive to tamoxifen (34) consistent with our results from a larger panel of ER+ cell lines. However, it is unclear how PIK3CA mutations contribute to the phenotype observed, namely of improved outcome with endocrine therapy.
Although the exact underlying mechanism is unclear, our data point to several possible hypotheses. The first is that this phenomenon could be mediated by unknown genes. Potential candidates are PP2A and/or PML, both known negative regulators of AKT1 and mTOR, and both present in the larger gene signature list (P < 0.05) (35, 36).
Second, considering that the PI3K pathway is known for its negative regulation abilities in the setting of chronic activation, mediated in part by reduction of IRS protein, we speculated whether feedback down-regulation was responsible for our observations (37, 38). Although we saw decreased IRS2 expression, classically the activation of the PI3K signaling that results in feedback down-regulation is a result of elevated mTOR/S6K signaling (39). Against this model, we saw low mTORC1 signaling in association with PIK3CA mutations in ER+/HER2− human BC. It is possible that another unknown feedback mechanism exists to down-regulate mTORC1 signaling. An interaction mediated by estrogen signaling is possible given the extensive cross-talk that exists between estrogen and growth factor pathways and could also contribute to increased tamoxifen sensitivity. However, typically activated PI3K/AKT is thought to contribute to the development of endocrine therapy resistance by augmentation of nuclear ER signaling (40).
The third potential explanation for our findings is that PIK3CA mutations are associated with weak pathway activation, i.e., the pathway is activated but other PI3K pathway alterations produce stronger pathway activation in BC. Viewed from this perspective, our observations could reflect “relatively low” activation as a result of the comparison containing some WT tumors with stronger downstream output (41). Therefore, it is possible that the good clinical outcome could still be mediated by classic feedback down-regulation secondary to pathway activation, albeit weak. This would then result in insensitivity to upstream receptor tyrosine kinase stimulation (37), which in turn would delay or prevent the development of growth factor receptor–mediated endocrine therapy resistance in PIK3CA or AKT1 mutated ER+/HER2− BC. This seems plausible considering that tamoxifen resistance is thought to be predominantly driven by growth factor receptor signaling (42).
It is conceivable that tumor aggressiveness in BC and the level of oncogenic signaling may vary depending on the mechanism of pathway activation. PIK3CA mutations in invasive endometrial cancer are reported not to be associated with an aggressive phenotype compared with tumors with decreased PTEN and PIK3CA amplification (43). Recent data in BC also support significant differences in functional PI3K output between PIK3CA mutations and PTEN loss (8, 29). The strong repressive abilities of intact PTEN may also contribute to a PIK3CA mutations having a “weaker” oncogenic effect. Moreover, KD mutation activation could be limited by p85 repression (44). The coexistence of multiple PI3K pathway aberrations in a single tumor supports the hypothesis that multiple “hits” may be required to induce effective tumorigensis as a result of the presence of strong repressors and/or to relieve the negative feedback on the pathway induced by one alteration (45).
Our study does not examine the effect of PIK3CA mutations in HER2+ or ER− disease; PIK3CA mutations are reported to increase trastuzumab resistance in HER2+ BC (46). It is also essential to emphasize that we did not develop the PIK3CA-GS to replace sequencing to determine those BCs that are PIK3CA mutated; rather, we demonstrate that these gene expression changes can provide insight into the functional steady state induced by a PIK3CA mutation in ER+/HER2− BC. Importantly, this seems not to be equivalent to that which is found in experimental models. Ultimately, the ability of the PIK3CA-GS to predict response to PI3K/mTOR inhibitors may be its most valuable application. Our cell line data provides some preliminary evidence of its potential clinical utility; however, this requires further validation. Nevertheless, our results from more than 1,500 human BCs suggest that, in future clinical trials conducted in ER+/HER2− BC, stratification for PIK3CA and AKT1 mutations will be important as these patients may respond well to hormonal therapy. This could also have implications for the clinical development of PI3K/mTOR inhibitors in BC.
Methods
Further information on methods is available in SI Methods.
PIK3CA and AKT1 Mutation Analysis.
DNA were obtained from 173 primary ER+ BC tumor samples from a previously described TAM dataset (Table S1) (47). Screening of mutations is described in SI Methods. Corresponding microarray data were used to develop the PIK3CA-GS. Microarray analyses were performed using BRB ArrayTools, version 3.5 (http://linus.nci.nih.gov/BRB-ArrayTools.html), and R software, version 2.3 (www.r-project.org). The PIK3CA-GS (Dataset S1) was a simple linear algorithm in which the weight of the genes was +1 or −1 depending on their association with PI3KCA mutation status: higher expression equates to the mt-like profile in ER+/HER2− human BC. Independent validation datasets of PIK3CA-GS's ability as a continuous variable to predict mutation status have been described for the MDACC (20) and Saal et al. (19) datasets.
Survival Analysis of PIK3CA-GS.
Samples analyzed were from women diagnosed with early-stage BC who had received adjuvant tamoxifen monotherapy [TAM dataset (n = 354), Table S1; ref. 47] or no systemic adjuvant treatment [UNT dataset (n = 1,189) as described in Desmedt et al. (48)]. BC molecular subtypes (ER−/HER2−, HER2+, and ER+/HER2−) were defined using a previously reported method (22, 48). The ER+/ HER2− low (L) and high (H) proliferative subgroups were defined as described in Loi et al. (16, 27), similar to the Luminal A and B subtypes described by Perou et al. (15)
BC Cell Lines.
The dataset described in Hu et al. (30) was used to evaluate the PIK3CA-GS's ability as a continuous variable to predict mutation status in ER+ (luminal) cell lines.
Senescence and Protein Evaluation.
For information on senescence and protein evaluation, see SI Methods.
Cell Culture and Proliferation Assays.
Half-maximal growth inhibitory concentrations (IC50) were assessed by using MTT proliferation assays.
Statistical analysis was performed using the SPSS statistical software package (SPSS), version 15.0. The performance of the predictive abilities of the PIK3CA-GS was evaluated by the AUC as a continuous variable. For Kaplan-Meier survival curves, the PIK3CA-GS was dichotomized to form two groups: the group with the higher expression of the PIK3CA-GS is referred to as mt-like, and the one with lower expression WT-like.
Supplementary Material
Acknowledgments
We thank N. Rosen for helpful discussion. S.L. is supported by the National Health and Medical Research Council of Australia (NHMRC) and the European Society of Medical Oncology. G.A.M. is supported by a Sir Edward Dunlop Fellowship of the Cancer Council of Victoria and the NHMRC. C.S. and B.H.K. are supported by the Breast Cancer Research Foundation, the MEDIC Foundation, and the Belgian National Foundation for Cancer Research.
Footnotes
Conflict of interest statement: A provisional worldwide patent was filed by Université Libre de Bruxelles for PIK3CA mutation gene signature: prognosis and therapeutic responsiveness of HER2 overexpressing and estrogen receptor–positive breast cancer. S.L., B.H.K., C.S., and G.A.M. are named inventors.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE6532 and GSE9195).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0907011107/-/DCSupplemental.
References
- 1.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 2.Isakoff SJ, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 3.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell IG, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 6.Levine DA, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 7.Saal LH, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 8.Stemke-Hale K, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbareschi M, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–6069. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 10.Lai YL, et al. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15:1064–1069. doi: 10.1245/s10434-007-9751-7. [DOI] [PubMed] [Google Scholar]
- 11.Li SY, Rong M, Grieu F, Iacopetta B. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–95. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- 12.Kalinsky K, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama N, et al. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res. 2007;13:408–414. doi: 10.1158/1078-0432.CCR-06-0267. [DOI] [PubMed] [Google Scholar]
- 14.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loi S, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 17.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 18.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liedtke C, et al. PIK3CA-activating mutations and chemotherapy sensitivity in stage II-III breast cancer. Breast Cancer Res. 2008;10:R27. doi: 10.1186/bcr1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpten JD, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 22.Wirapati P, et al. Meta-analysis of gene expression profiles in breast cancer: Toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkegaard T, et al. AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J Pathol. 2005;207:139–146. doi: 10.1002/path.1829. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol Cell Biol. 2007;27:662–677. doi: 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sotiriou C, et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 28.Creighton CJ. A gene transcription signature of the Akt/mTOR pathway in clinical breast tumors. Oncogene. 2007;26:4648–4655. doi: 10.1038/sj.onc.1210245. [DOI] [PubMed] [Google Scholar]
- 29.Vasudevan KM, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X, et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 31.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 32.Di Cosimo S, Baselga J. Phosphoinositide 3-kinase mutations in breast cancer: A “good” activating mutation? Clin Cancer Res. 2009;15:5017–5019. doi: 10.1158/1078-0432.CCR-09-1173. [DOI] [PubMed] [Google Scholar]
- 33.Ellis MJ, et al. Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2010;119:379–390. doi: 10.1007/s10549-009-0575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whyte DB, Holbeck SL. Correlation of PIK3Ca mutations with gene expression and drug sensitivity in NCI-60 cell lines. Biochem Biophys Res Commun. 2006;340:469–475. doi: 10.1016/j.bbrc.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 35.Trotman LC, et al. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441:523–527. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernardi R, et al. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- 37.Paz K, et al. A molecular basis for insulin resistance. Elevated serine/threonine phosphorylation of IRS-1 and IRS-2 inhibits their binding to the juxtamembrane region of the insulin receptor and impairs their ability to undergo insulin-induced tyrosine phosphorylation. J Biol Chem. 1997;272:29911–29918. doi: 10.1074/jbc.272.47.29911. [DOI] [PubMed] [Google Scholar]
- 38.Li J, DeFea K, Roth RA. Modulation of insulin receptor substrate-1 tyrosine phosphorylation by an Akt/phosphatidylinositol 3-kinase pathway. J Biol Chem. 1999;274:9351–9356. doi: 10.1074/jbc.274.14.9351. [DOI] [PubMed] [Google Scholar]
- 39.O'Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborne CK, Shou J, Massarweh S, Schiff R. Crosstalk between estrogen receptor and growth factor receptor pathways as a cause for endocrine therapy resistance in breast cancer. Clin Cancer Res. 2005;11:865s–870s. [PubMed] [Google Scholar]
- 41.Loi S, et al. Gene expression profiling identifies activated growth factor signaling in poor prognosis (Luminal-B) estrogen receptor positive breast cancer. BMC Med Genomics. 2009;2:37. doi: 10.1186/1755-8794-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massarweh S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 43.Salvesen HB, et al. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc Natl Acad Sci USA. 2009;106:4834–4839. doi: 10.1073/pnas.0806514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: Variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junttila TT, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 47.Loi S, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desmedt C, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.