Abstract
Inhibition of natural killer (NK) cells is mediated by MHC class I receptors including the killer cell Ig-like receptor (KIR). We demonstrate that HLA-C binding peptides can function as altered peptide ligands for KIR and antagonize the inhibition mediated by KIR2DL2/KIR2DL3. Antagonistic peptides promote clustering of KIR at the interface of effector and target cells, but do not result in inhibition of NK cells. Our data show that, as for T cells, small changes in the peptide content of MHC class I can regulate NK cell activity.
Keywords: killer cell immunoglobulin-like receptors, MHC class I
Natural killer (NK) cells have a fundamental role in the immune response to tumors and viruses. The inhibitory receptors for MHC class I regulate both target cell recognition and NK cell education, and include the killer cell Ig-like receptors (KIRs) that recognize classical HLA-A, -B, and -C ligands. The specificity of inhibitory KIR for MHC is determined by motifs on the MHC class I heavy chain, for example KIR2DL2 and KIR2DL3 bind HLA-C allotypes with asparagine at position 80 of the heavy chain. However, the interaction of KIR with their cognate MHC class I ligands can be modulated by the bound peptide (1–5). In particular, residues 7 and 8 of the peptide bound to MHC class I can promote or abrogate binding of KIR. Despite the extremely rapid evolution of the KIR gene family, all inhibitory KIRs tested to date have retained the feature of peptide selectivity (6–11).
Recognition of target cells by NK cells is determined by the integration of the signals derived from both activating and inhibitory receptors. Thus, attenuation of an inhibitory signal can lead to enhanced activation of NK cells. This has been related to wholesale changes in MHC class I and viruses, and tumors that do not induce MHC class I down-regulation may therefore be considered to be relatively resistant to NK cells attack (12). During viral infections and tumorigenesis, the MHC class I peptide repertoire changes so that viral epitopes or tumor antigens can be presented to cytotoxic effector T cells. Peptide elution studies have shown that viral infections can change the repertoire of self and viral peptides presented by MHC class I (13, 14). Therefore, recognition of changes in peptide repertoire by the innate immune system may confer a selective advantage to the host.
Changes in endogenously derived peptides may perturb NK cell inhibition mediated by KIR3DL1 and also the CD94:NKG2A heterodimer in a manner that is poorly understood (15, 16). One model is that peptide:MHC complexes that do not bind inhibitory receptors strongly play a purely passive role in the process and are ignored by the NK cell. Alternatively, they may play a more active role in target cell recognition. As changes in peptide repertoire represent a potentially important mechanism for enhanced recognition of NK cell targets, we have investigated their potential to affect NK cell inhibition by KIR. Our results suggest that peptides that bind MHC class I but not KIR can act as altered peptide ligands and efficiently reduce KIR-mediated inhibition in a manner consistent with peptide antagonism.
Results
KIR Binding of Single Peptides to HLA-C: Peptide Correlates with Inhibition.
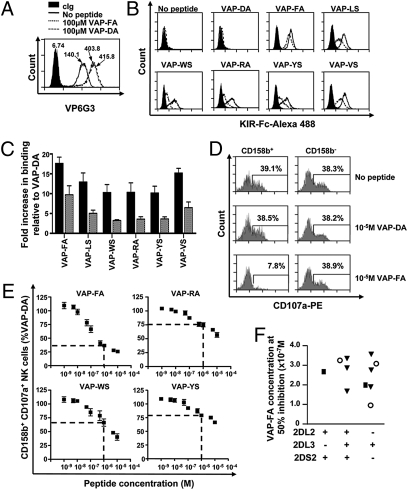
We screened peptide variants that confer binding of KIR2DL2 and KIR2DL3 to HLA class I, using the TAP-deficient T2 cell line. In the absence of exogenous peptide, the TAP-deficient T2 cell line expresses HLA-Cw*0102 on the cell surface as detected by the Cw*0102 specific Ab VP6G3 (17) (Fig. 1 and Fig. S1A). Cell surface MHC-I on T2 cells contains low-affinity peptide (18), which most likely dissociates rapidly and hence does not inhibit NK cells (Fig. S1B). HLA-Cw*0102 can therefore be stabilized with exogenous peptide (Fig. 1A). We screened peptide variants based on the nonamer peptide VAPWNSLSL for binding of KIR2DL2 and KIR2DL3 to HLA-Cw*0102. VAPWNSLSL was the most abundant identified peptide eluted from HLA-Cw*0102 expressing 721.221 cells and is derived from the TIMP1 protein (19). The VAPWNSLSL derivatives retained the anchor residues for MHC class I binding, but varied at P7 and P8, which is predicted to affect KIR binding (10, 19, 20). The 58 derivatives synthesized had leucine at P7 with all potential residues at P8 and either alanine or serine at P8 with all potential residues at P7. Stabilization of Cw*0102 was confirmed by VP6G3 and KIR binding was detected with KIR-Fc fusion constructs of KIR2DL2 and KIR2DL3 (Table S1). In the absence of peptide, there was no significant binding of KIR2DL2-Fc and KIR2DL3-Fc to HLA-Cw*0102. Twenty-four peptides caused greater than twofold binding of KIR2DL2 to HLA-Cw*0102 and eight caused greater than twofold binding of KIR2DL3 to HLA-Cw*0102 compared with no peptide. As previously described, there was a preference for large amino acids with high hydrophobicity at P7 and a small amino acid at P8 (10, 21, 22). Seven peptides that induced a range of KIR binding to HLA-Cw*0102 were studied further. These included VAPWNSLSL (VAP-LS), VAPWNSDAL (VAP-DA), VAPWNSFAL (VAP-FA), VAPWNSWSL (VAP-WS), VAPWNSRAL (VAP-RA), VAPWNSYSL (VAP-YS), and VAPWNSVSL (VAP-VS; Fig. 1B). At a concentration of 100 μM these peptides stabilized HLA-Cw*0102 to similar levels (Fig. S2). The avidity of KIR for HLA-Cw*0102 was influenced by different peptides and the hierarchy for KIR2DL2, or KIR2DL3, binding to HLA-Cw*0102:peptide complexes was determined as VAP-FA > VAP-VS ≈ VAP-LS > VAP-WS ≈ VAP-YS ≈ VAP-RA (Fig. 1C and Fig. S3). There was no discernible binding of KIR2DL2-Fc and KIR2DL3-Fc to HLA-Cw*0102:VAP-DA. With the exception of VAP-DA, KIR2DL2-Fc bound to HLA-Cw*0102: peptide significantly more avidly than KIR2DL3-Fc (P < 0.05; Mann-Whitney U test), but both KIRs had similar peptide hierarchies (Fig. 1C and Fig. S3). The strong binding peptide VAP-FA, and three weaker binding peptides, VAP-RA, VAP-WS, and VAP-YS, were incorporated into CD107a degranulation assays. The VAP-FA peptide promoted strong binding of both KIR2DL2 and KIR2DL3 to HLA-Cw*0102, and strongly inhibited degranulation of CD158b+ NK cells, but not CD158b− NK cells (Fig. 1D). The VAP-DA peptide did not induce binding of KIR-Fc to HLA-Cw*0102 and did not inhibit degranulation of CD158b+ NK cells. The peptide hierarchy in CD017a assays were similar to those of KIR-Fc binding assays (Fig. 1E). To determine the effect of KIR and HLA genotype on the level of inhibition, we tested 11 individuals with different KIR2DL2/KIR2DL3/KIR2DS2 and HLA-C types and calculated the values for 50% inhibition by VAP-FA for each one (Fig. 1F and Table S2). These did not differ significantly among individuals with different KIR genotypes (P > 0.05; one-way ANOVA).
Fig. 1.
Comparison of KIR binding and NK cell inhibition for peptide variants. (A) Stabilization of HLA-Cw*0102 on T2 cells as determined by VP6G3 in the absence of peptide or with peptides VAP-FA and VAP-DA at 100 μM as determined by flow cytometry. MFI values are shown. (B) Flow cytometry histograms of the binding of KIR-Fc to T2 cells in the absence or presence of 100 μM of the indicated peptides. Cells were stained with 60 μg/mL isotype control (KIR2DS4-Fc; filled histogram), KIR2DL2-Fc (solid line), or KIR2DL3-Fc (dashed line) fusion proteins conjugated to protein-A Alexa-488. (C) The relative increase, as determined by flow cytometry, in KIR2DL2-Fc (black bar) or KIR2DL3-Fc (gray bar) binding to HLA-Cw*0102 stabilized with 100 μM of the indicated peptides compared with 100 μM VAP-DA. KIR-Fc was used at 60 μg/mL The means ± SEM of three independent experiments are shown. (D) Degranulation of NK cells in response to T2 cells in the absence or presence of 10 μM indicated peptide. Histogram plots are gated on CD3−CD56+CD158b+ or CD3−CD56+CD158b− NK cells. The frequency of degranulating NK cells is shown for each subset. (E) CD3−CD56+CD158b+ NK cell degranulation to T2 targets incubated with increasing concentrations of the indicated peptides. Data are normalized to degranulation in response to T2 cells incubated with VAP-DA and the level of inhibition at 1 μM peptide is indicated by a dashed line. No inhibition of CD3−CD56+CD158b− NK cells was observed. The percentage of CD158b+CD107a+ NK cells normalized to VAP-DA and the means ± SEM of three independent experiments are shown. (F) The peptide concentration required to inhibit 50% NK cells relative to VAP-DA for 11 donors with different KIR haplotypes. Squares indicate individuals with two group 2 HLA-C allotypes; circles, two group 1 HLA-C allotypes; and triangles, one group 1 and one group 2 HLA-C allotype.
Peptides Inducing Weak KIR Binding Antagonize NK Cell Inhibition.
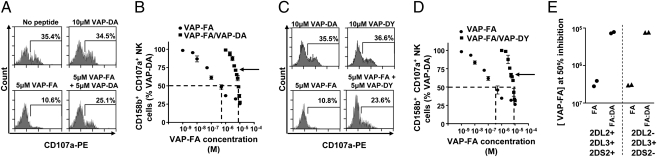
The cell surface expression level of HLA-Cw*0102 on T2 cells is maximal at approximately 10 μM of exogenous peptide (Fig. S2B). To compare the effects of changing MHC class I expression with that of changing peptide repertoire, we used combinations of peptides that confer strong and weak recognition of HLA-Cw*0102 to KIR. Different ratios of VAP-FA and VAP-DA, varying by 10% increments, were used to stabilize HLA-Cw*0102 on T2 cells, and the results obtained were compared with those achieved by using VAP-FA alone. The peptide concentration of VAP-FA was titrated up to a maximum of 10 μM, whereas the final peptide concentration of all of the VAP-FA:VAP-DA mixes was kept constant at 10 μM. HLA-Cw*0102 levels were similar in all peptide mixes (Fig. S4). The peptide-loaded T2 cells were used as target cells in CD107a assays (Fig. 2). The inhibition of CD158b+ NK cells by the VAP-FA peptide was significantly reduced in the presence of the VAP-DA peptide compared with VAP-FA alone at all concentrations [P < 0.001; analysis of covariance (ANCOVA); Fig. 2 A and B]. For instance, at 5 μM VAP-FA, we observed degranulation of 10.6% of CD158b+ NK cells in the absence of VAP-DA, but 25.1% in its presence at 5 μM. To exclude the possibility that this effect was a result of VAP-DA having a faster on-rate for MHC I than VAP-FA, we added the peptides to the T2 cells sequentially. No significant difference in degranulation of CD158b+ NK cells was observed if VAP-FA was added before VAP-DA compared with adding VAP-FA after VAP-DA (Fig. S5)
Fig. 2.
A weak KIR-binding peptide can disrupt inhibition by a strong KIR-binding peptide. (A) Degranulation of CD3−CD56+CD158b+ NK cells in response to T2 cells alone or incubated with VAP-DA and VAP-FA alone or in combination. (B) The fraction of degranulating CD3−CD56+CD158b+ NK cells in response to T2 cells incubated with different concentrations of VAP-FA alone or in combination with VAP-DA. Degranulation of CD3−CD56+CD158b+ NK cells in response to VAP-FA and VAP-DY alone or in combination shown as flow cytometry plots (C) or titrations (D). For the peptide mix experiments, different ratios of VAP-FA and VAP-DA or VAP-FA and VAP-DY starting at 10% VAP-FA/90% VAP-DA or 10% VAP-FA/90% VAP-DY, finishing at 100% VAP-FA, and varying by 10%, are shown. The final total peptide concentration in these mixes was fixed at 10 μM. Data are normalized to the degranulation in response to T2 cells incubated with 10 μM VAP-DA, and the dashed line indicates the concentration of VAP-FA at which 50% of NK cells are inhibited relative to VAP-DA. The means ± SEM of three independent experiments are shown for each peptide. (E) Concentration of VAP-FA at which 50% inhibition values were obtained for four donors with different KIR genotypes. Shown are the values for VAP-FA alone (FA) or for VAP-FA and VAP-DA (FA:DA).
VAP-DA has a residue at P8 that is permissive for KIR binding, but tyrosine at P8 is not (10) (Table S1). A second KIR-nonbinding peptide VAPWSNDYL (VAP-DY) was synthesized. CD158b+ NK cell inhibition mediated by VAP-FA was dramatically reduced in the presence of a VAP-DY (P < 0.001; ANCOVA; Figs. 2 C and D). Thus, peptides that in isolation do not induce target cell recognition by NK cells can significantly depress the inhibition as a result of a strong KIR binding peptide and so act as altered peptide ligands.
Presence of KIR2DS2 Does Not Affect Peptide Antagonism.
The levels of inhibition of CD158b+ NK cells from two KIR2DL2−KIR2DS2−KIR2DL3+ and two KIR2DL2+KIR2DS2+KIR2DL3+ donors were compared (Fig. 2E). Overall half-maximal inhibition of CD158b+ NK cells by VAP-FA required 20 times higher peptide concentration in the presence of VAP-DA than in its absence for all donors. No significant differences in 50% inhibition values for VAP-FA in the presence or absence of VAP-DA were observed between donors expressing either KIR genotype (P > 0.05; one-way ANOVA), implying that KIR2DS2 does not contribute to the observed results, and that VAP-DA specifically perturbs inhibitory sig-nals generated by the interaction of KIR2DL2/KIR2DL3 with HLA-Cw*0102:VAP-FA. Consistent with this, we observed no binding of KIR2DS2 to any of the peptides assayed in our screen (Table S1).
Antagonism Is Determined by the Relative Concentrations of Peptides Inducing Strong or Weak KIR Binding.
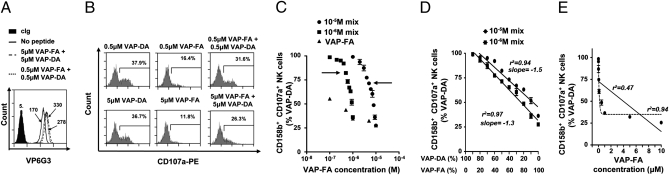
To test if this change in inhibition is a result of VAP-DA outcompeting VAP-FA for HLA-C, we performed further CD107a assays at 1 μM total peptide concentration. There were consistently lower levels of both HLA-C expression and inhibition at 1 μM VAP-FA than at 10 μM (Fig. 1E and Fig. S2B). Therefore, when the total peptide concentration is 1 μM, free HLA-Cw*0102 was available for loading by any VAP-FA that might have been competed out by VAP-DA. At a 50:50 ratio of VAP-FA:VAP-DA at either 1 μM or 10 μM peptide concentration, increased activation of CD158b+ NK cells was observed compared with the same concentration for T2 cells loaded with VAP-FA alone (Fig. 3 A–C). The slopes of the titration curves were similar at both peptide concentrations (P > 0.05; ANCOVA; Fig. 3D), indicating that the change of inhibition observed in the presence of VAP-DA is not caused by VAP-DA competing out VAP-FA, but dependent on the ratio of strong to weak KIR-binding peptides. Furthermore, regression analysis showed that the VAP-FA:VAP-DA peptide mix gave a linear change in activation (r2 = 0.94), whereas the titration of FA fits a single exponential (r2 = 0.94; Fig. 3E). Thus, the change in inhibition induced by changing the concentration of VAP-FA has a distinct kinetic from that induced by changing the ratio between VAP-FA and VAP-DA.
Fig. 3.
The ratio of strong to weak KIR-binding peptide determines the change in inhibition. (A) Stabilization of HLA-Cw*0102 on T2 cells in the absence or presence of 1 μM or 10 μM peptide. (B) Degranulation of CD3−CD56+CD158b+ NK cells in response to T2 cells alone or incubated with a single peptide or a combination of VAP-DA and VAP-FA at 1 μM or 10 μM final. (C) Fraction of degranulating CD3−CD56+CD158b+ NK cells in response to T2 cells incubated with VAP-FA alone or in combination with VAP-DA. The peptide mixes tested consisted of different ratios of VAP-FA and VAP-DA, from 10% VAP-FA/90% VAP-DA to 100% VAP-FA, and differing by 10%, to a final total peptide concentration of 1 μM or 10 μM. Data are normalized to the degranulation observed in response to T2 cells incubated with 10 μM VAP-DA and represent the means ± SEM of three independent experiments. Arrows indicate the 50% VAP-FA point. (D) shows a linear regression of the data from C plotted as the fraction of degranulating CD3−CD56+CD158b+ NK cells against the ratio of VAP-FA to VAP-DA in the peptide mix. (E) Regression analyses for the peptide titration of VAP-FA alone. The full line indicates a linear regression and the dashed line a one-phase decay nonlinear regression. Correlation coefficients for both analyses are shown.
A Peptide That Induces Intermediate Binding of KIR to MHC Does Not Antagonize the Inhibition as a Result of a Peptide Inducing Strong Binding.
The intermediate binding peptide VAP-RA induces a lower level of NK cell inhibition than VAP-FA. To determine if this affects the potential for antagonism, we performed T2 assays using mixes of VAP-RA and VAP-DA made to a final concentration of 10 μM (Fig. 4 A and B). Inhibition by VAP-RA is markedly perturbed by VAP-DA such that, at a ratio of 60% VAP-RA to 40% VAP-DA, a minimal inhibition of NK cells was observed (Fig. 4B). This compared with a ratio of 10% VAP-FA to 90% VAP-DA (Fig. 2B). Thus, an intermediate KIR-binding peptide is more efficiently antagonized than a strong KIR-binding one.
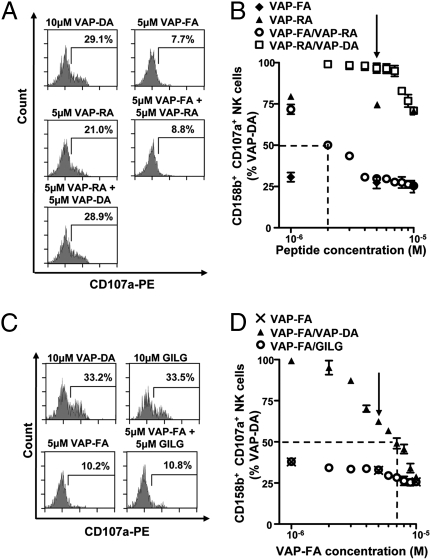
Fig. 4.
Intermediate KIR-binding or HLA-A2 binding peptides do not antagonize inhibition by a strong KIR-binding peptide. T2 cells were incubated with VAP-FA, VAP-RA, or VAP-DA alone or in combination and degranulation assays performed. Flow cytometry plots (A) and the results of peptide mix titrations (B) are shown. Peptide mixes were made to 10 μM final concentration. For the titration of VAP-FA and the VAP-FA:VAP-RA combination, the concentration of VAP-FA is plotted, and for VAP-RA and VAP-RA:VAP-DA, that of VAP-RA is plotted. The dashed line indicates the concentration of VAP-FA at which 50% of NK cells relative to VAP-DA alone are inhibited for the VAP-FA:VAP-RA mix. The arrow indicates the 50:50 peptide mix point. (C and D) T2 cells were incubated with VAP-FA or an HLA-A*0201-binding peptide (GILG). (C) Degranulation of CD3−CD56+CD158b+ NK cells in response to T2 cells alone or incubated with the indicated peptides. (D) The fraction of degranulating CD3−CD56+CD158b+ NK cells in response to T2 cells incubated with VAP-FA alone or in combination with GILG or VAP-DA to a final concentration of 10 μM. The dashed line indicates the concentration of VAP-FA at which 50% of NK cells relative to VAP-DA alone are inhibited for the VAP-FA:VAP-DA mix. The arrow indicates the 50:50 peptide mix point. For all experiments, data are normalized to the degranulation observed in response to T2 cells incubated with 10 μM VAP-DA, and the means ± SEM of three independent experiments are shown.
To determine if a peptide inducing intermediate binding could also antagonize one inducing strong binding, we tested peptide mixes of VAP-FA and VAP-RA (Fig. 4 A and B). There was a minimal reduction in inhibition at ratios of VAP-FA:VAP-RA up to 60%:40%. Thus, VAP-RA induces low levels of NK cell inhibition but it does not antagonize VAP-FA–mediated inhibition. Thus, antagonism is a feature of peptides that do not inhibit NK cells as distinct from peptides that can induce low-level inhibition of NK cells. This implies that there is a threshold of binding that discriminates peptide:MHC class I complexes with weak affinity for KIR from antagonistic ones.
Peptide Antagonism Is Specific for Cognate MHC.
T2 cells also express HLA-A2, and therefore to determine whether the generic up-regulation of HLA class I can antagonize KIR-mediated inhibition, we loaded T2 cells with VAP-FA in the presence of an HLA-A*0201–specific influenza matrix-derived peptide, GILGFVFTL (GILG) (23). GILG did not up-regulate HLA-Cw*0102, and similarly VAP-FA did not significantly up-regulate HLA-A*0201 (Fig. S6). The peptide GILG did not disrupt CD158b+ NK cell inhibition mediated by VAP-FA (Fig. 4 C and D). This indicates that the disruption of inhibition by KIR2DL2/KIR2DL3:HLA-C requires the presence of a cognate MHC ligand.
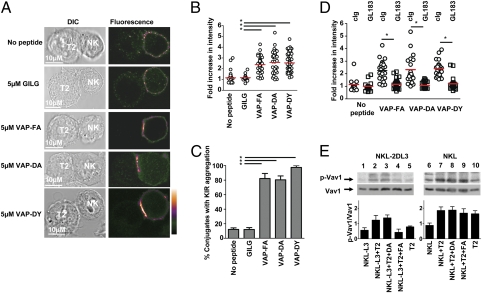
An Antagonist Peptide Induces Clustering of KIR at the Interface Between NK Cells and Target Cells, but Not Dephosphorylation of Vav1.
As there was a requirement for cognate MHC to perturb inhibition of NK cells by a strong KIR-binding peptide, we investigated whether HLA-Cw*0102:VAP-DA could induce aggregation of KIR at the interface between the effector and target cells. NKL cells transfected with KIR2DL3 (NKL-KIR2DL3) were imaged for KIR aggregation following coincubation with T2 cells that were unloaded or loaded with a control peptide (GILG), VAP-FA, VAP-DA, or VAP-DY (Fig. 5A). Quantification of the intensity of KIR2DL3 at the interface between effector and target cells showed that T2 cells loaded with VAP-DA or VAP-DY alone leads to KIR accumulation to the same extent as VAP-FA alone and induces a similar percentage of conjugates with KIR2DL3 aggregation (Fig. 5 B and C). This clustering was efficiently blocked by the anti-KIR2DL3 antibody GL183 (Fig. 5D). Thus, antagonist peptides can induce aggregation of KIR, but additional receptor ligand interactions may be required to facilitate this (24). Furthermore, although VAP-DA induced KIR clustering, it did not induce dephosphorylation of the SHP-1 substrate Vav1 compared with unloaded T2 cells (Fig. 5E). Additionally VAP-FA, but not VAP-DA, inhibited polarization of the microtubule organizing center (Fig. S7). Thus, antagonist peptides facilitate KIR aggregation but uncouple this process from inhibitory signaling.
Fig. 5.
The weak KIR-binding peptide VAP-DA mediates KIR clustering at the interface between NK cells and target cells. (A) Clustering at the interface between NKL-2DL3 cells and T2 targets in the absence of peptide or in the presence of the indicated peptides. Images are labeled with a pseudocolor scale. (B) The increase in fluorescence intensity at the interface between NKL-2DL3 and the T2 cells compared with the NKL plasma membrane at a noncontact area (means are indicated in red). (C) The percentage of conjugates with KIR clustering depicted as the means and SDs from three independent experiments. A minimum of 17 conjugates were counted per condition per experiment. (D) The effect of addition of GL183 on clustering between peptide pulsed T2 cells and NKL-2DL3. In all panels, an asterisk indicates a significant difference of P < 0.0001 (Student t test). (E) Western blot analysis of KIR2DL3 transfected NKL (Left) or untransfected NKL (Right) cells incubated with T2 cells unloaded (lanes 2 and 7) or loaded overnight with the peptides VAP-DA (lanes 3 and 8) and VAP-FA (lanes 4 and 9) at 20 μM and then analyzed for the presence of absence of VAV1 and phosphorylated VAV1. Baseline signals from NKL-2DL3 (lane 1), NKL-2DL3 (lane 6), and T2 cells (lanes 5 and 10) are also shown. The mean quantitation of the ratio of pVAV1 to VAV1 from three independent experiments ± SEM is shown graphically.
Discussion
We show that KIR-positive NK cells respond more readily to changes in peptide than to changes in MHC class I expression. Tumors and viruses may not down-regulate MHC class I substantially, but may change peptide repertoire, and so peptide selectivity confers NK cells with an additional sensitive recognition mechanism (1). We have used a reductionist system to explore how changes in MHC class I peptide repertoire may influence NK cell recognition. The economics of this system are such that, at a ratio of 5 μM KIR-binding peptide (VAP-FA) to 5 μM weak KIR-binding peptide (VAP-DA/DY), the number of NK cells degranulating was the same as for 0.1 μM KIR-binding peptide alone. This indicates the antagonistic effect of peptides that induce weak KIR binding to MHC class I and so act as altered peptide ligands for KIR. In vivo MHC class I presents a wide array of peptides with different potentials to induce binding of KIR to MHC class I. Our work has distinguished at least three different types of peptide in this context: those inducing strong inhibition (VAP-FA), those inducing low-level inhibition (VAP-RA), and antagonist peptides (VAP-DA). The ability of changes in peptide repertoire to alter NK cell recognition in vivo will depend on the relative amounts of these types of peptides, and also the nature of the peptides produced by infection or tumorigenesis. Viral peptides can dominate the peptide repertoire after infection (25), and this may facilitate recognition of an infected target. Alternatively, a viral peptide may induce strong inhibitory KIR binding and so escape from NK cell recognition. The physiological relevance of our findings will require further analysis to determine the “set point” of the peptide repertoire of specific MHC class I allotypes for KIR recognition and how this may be induced to change.
Our observations are similar to “TCR antagonism” in which small quantities of antagonist peptides perturb CD8+ T cell activation by cognate peptide (26–28). Antagonism by VAP-DA/DY is distinct from the findings with the intermediate KIR binding peptide VAP-RA. Thus, KIR antagonism is distinct from low-level inhibition, and one feature of antagonistic peptides is that they are unable to induce inhibition. The second feature of an antagonistic peptide is that it uncouples KIR clustering from inhibitory signaling.
Inhibitory signaling in NK cells requires the formation of microclusters of signaling molecules (29). There were different kinetics of inhibition between decreasing VAP-FA concentration and changing the VAP-FA:VAP-DA ratio, implying that distinct mechanisms are operating to prevent formation of a productive inhibitory signal. One explanation for this may be that decreasing VAP-FA concentrations reduces the number of inhibitory microclusters, whereas the antagonist peptide primarily prevents their formation.
We did not detect binding of KIR2DL2 or KIR2DL3 to T2 cells loaded with VAP-DA. However, it has recently been shown that the TCR-peptide:MHC interaction has an increased affinity when measured in situ compared with in solution (30). This may be a result of signaling within the effector cell, the restriction imposed by the intercellular volume, or lateral interactions (31). These effects may explain why peptides that do not induce detectable binding of KIR to MHC in solution can still promote KIR clustering. We also observed a higher avidity of KIR2DL2 compared with KIR2DL3 for HLA-Cw*0102 and peptide. This increased avidity is a result of polymorphisms that change the hinge angle between the D1 and D2 domains of KIR2DL2 and KIR2DL3, rather than polymorphisms at MHC class I contact residues, and may account for the apparent differences in specificity of KIR2DL2 and KIR2DL3 (21). Consistent with this, we found that the peptide hierarchies of KIR2DL2 and KIR2DL3 closely overlap. Nevertheless, in our peptide screen, there were more KIR2DL3 nonbinders than KIR2DL2 nonbinders. KIR2DL3-positive NK cells may therefore be sensitive to a wider spectrum of changes in peptide repertoire than KIR2DL2-positive NK cells as a greater fraction of peptides could be antagonistic. This may in part explain the genetic association of KIR2DL3, but not KIR2DL2, with protection from hepatitis C infection (32, 33).
Loss of inhibition may be a result of a change in the self-peptides bound to HLA class I as opposed to a specific viral or tumor-derived peptide. Furthermore, MHC class I peptides may also be derived from the defective ribosomal products of newly synthesized proteins (34, 35). Changes in defective ribosomal products can occur very early, possibly within 45 min following infection, and peptide elution experiments have documented changes at 2 h (14, 36, 37). Thus, recognition of these early events of viral infection by NK cells could have a selective advantage for the host as evidenced by the maintenance of peptide selectivity by the inhibitory KIR.
NK cells function by integrating signals from activating and inhibitory receptors. The inhibitory receptors have an essential role in NK cell education and target cell recognition. Whereas class I down-regulation is one model for recognition of abnormal cells by NK cells, we propose that changing the MHC class I peptide repertoire represents a much more rapid and potent mechanism for removing dominant inhibitory signals, thus leading to NK cell activation.
Methods
Cell Lines and Culture.
The following cell lines were used: T2 (38), NKL, or NKL cells expressing KIR2DL3 (21). T2 cells were cultured in R10 medium [RPMI medium 1640 supplemented with 1% penicillin streptomycin (Invitrogen) and 10% FCS (Lonza)]. NKL and NKL-KIR2DL3 were cultured in R10 medium with 500 IU recombinant IL-2 (National Cancer Institute Biometric Research Branch; Fisher BioServices).
Antibodies.
Details of the sources of the antibodies used are given in SI Methods.
KIR Binding Assay.
The KIR-Fc fusion proteins KIR2DL2-Fc, KIR2DL3-Fc, KIR2DS2-Fc,and KIR2DS4-Fc were purified as described (39). KIR-Fc integrity was verified by ELISA using the antibody GL183 (Abcam). T2 cells (1 × 105) were incubated with peptide overnight at 26 °C. KIR-Fc proteins were conjugated with protein-A Alexa 488 (Invitrogen) at a molar ratio of 6:1 KIR-Fc:Prot-A. Peptide-pulsed T2 cells were then stained with KIR-Fc for 1 h at room temperature or with VP6G3 (17) followed by FITC-conjugated goat anti-human IgM antibody (Serotec).
Peripheral Blood Mononuclear Cell Isolation and Genotyping.
Peripheral blood mononuclear cells (PBMCs) were isolated using Hypaque-Ficoll (Amersham Biosciences) density centrifugation. HLA-C typing was performed by sequence specific oligonucleotide hybridization. KIR typing was performed using the method of Uhrberg et al. (40).
NK Cell Degranulation Assay.
PBMCs were stimulated overnight with 1 ng/mL rHuIL-15 (R&D Systems). T2 cells (4 × 104) were incubated with peptide at 26 °C overnight, washed, and resuspended with the PBMCs at an effector-to-target (E:T) ratio of 5:1 in fresh R10 medium containing peptide and anti–CD107a-PE or control IgG1-PE at 20 μL/mL. Cells were incubated at 26 °C for 4 h with 6 μg/mL Golgi-Stop (BD Biosciences) added after 1 h. Cells were stained and analyzed on a CyAn ADP Analyzer (Beckman Coulter) with Summit version 4.3 (Dako).
Western Blotting.
T2 cells were cultured with 20 μM peptide overnight at 26 °C. NK cell lines and T2 cells were coincubated at a 1:1 E:T ratio for 5 min at 37 °C. Cells were lysed in 20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM EDTA, 1 mM sodium orthovanadate, and 0.5% Triton X-100 and analyzed by Western blotting. Membranes were stripped using the Western Blot Recycling Kit (Alpha Diagnostics). Protein bands were detected by chemiluminescence (Supersignal Westpico Chemiluminescent Substrate; Perbio Science) using the ChemiDoc-It Imaging System and Vision Works software (UVP) and quantified with ImageJ software (National Institutes of Health).
Microscopy.
T2 cells were incubated overnight at 26 °C in with 10 μM peptide, then coincubated with NKL cells at an E:T ratio of 2:1 for 10 min at 37 °C. Cells were fixed in 2% paraformaldehyde for 0.5 h at 37 °C before permeabilizing with 0.01% Triton X-100 and staining with 20 μg/mL GL183 and Alexa Fluor-488 goat anti-mouse IgG. Cells were imaged using a Leica SP5 resonance scanning microscope (Leica Microsystems). Transmission images and FITC emission were collected in separate channels. Data were processed using Leica imaging software (Leica Microsystems) and ImageJ. The increase in fluorescence intensity at the immune synapse was calculated as a ratio of the average fluorescence intensity along the NKL-T2 interface compared with the average fluorescence intensity along the NKL plasma membrane not in contact with another cell, with both values corrected for background fluorescence as measured within an empty region of the image. For the KIR2DL3 blocking experiment, NKL-KIR2DL3 was incubated with 20 μg/mL GL183 mAb for 30 min at 4 °C before addition of T2 cells and cocultured at 37 °C for 10 min. Then cells were permeabilized, stained, and analyzed as described earlier.
Peptide Synthesis.
The peptide library was purchased from Mimotopes. All other peptides were purchased from Peptide Protein Research. Their identity was confirmed by MS and purity was greater than 95%.
Statistical Analysis.
All statistical analyses were performed using GraphPad Prism, version 5 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Dr. E. Long (NIAID) for the KIR-Fc fusion constructs, Dr. P. Parham (Stanford University) for the NKL-2DL3 cell line and comments on the manuscript, and The Facility of Imaging by Light Microscopy for assistance with microscopy. This work was supported by fellowships from the Medical Research Council, United Kingdom (to L.F.), and The Wellcome Trust, United Kingdom (to S.I.K.). T.E. was supported by Cancer Research UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913745107/-/DCSupplemental.
References
- 1.Malnati MS, et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 2.Zappacosta F, Borrego F, Brooks AG, Parker KC, Coligan JE. Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc Natl Acad Sci USA. 1997;94:6313–6318. doi: 10.1073/pnas.94.12.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maenaka K, et al. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J Biol Chem. 1999;274:28329–28334. doi: 10.1074/jbc.274.40.28329. [DOI] [PubMed] [Google Scholar]
- 4.Stewart CA, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci USA. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thananchai H, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valés-Gómez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci USA. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peruzzi M, Wagtmann N, Long EO. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med. 1996;184:1585–1590. doi: 10.1084/jem.184.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansasuta P, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 10.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 11.Mandelboim O, Wilson SB, Valés-Gómez M, Reyburn HT, Strominger JL. Self and viral peptides can initiate lysis by autologous natural killer cells. Proc Natl Acad Sci USA. 1997;94:4604–4609. doi: 10.1073/pnas.94.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobigs M, Müllbacher A, Regner M. MHC class I up-regulation by flaviviruses: Immune interaction with unknown advantage to host or pathogen. Immunol Cell Biol. 2003;81:217–223. doi: 10.1046/j.1440-1711.2003.01161.x. [DOI] [PubMed] [Google Scholar]
- 13.Hickman HD, et al. Cutting edge: Class I presentation of host peptides following HIV infection. J Immunol. 2003;171:22–26. doi: 10.4049/jimmunol.171.1.22. [DOI] [PubMed] [Google Scholar]
- 14.Meiring HD, et al. Stable isotope tagging of epitopes: A highly selective strategy for the identification of major histocompatibility complex class I-associated peptides induced upon viral infection. Mol Cell Proteomics. 2006;5:902–913. doi: 10.1074/mcp.T500014-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Liberatore C, et al. Natural killer cell-mediated lysis of autologous cells modified by gene therapy. J Exp Med. 1999;189:1855–1862. doi: 10.1084/jem.189.12.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaëlsson J, et al. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J Exp Med. 2002;196:1403–1414. doi: 10.1084/jem.20020797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulder A, et al. A human monoclonal antibody against HLA-Cw1 and a human monoclonal antibody against an HLA-A locus determinant derived from a single uniparous female. Tissue Antigens. 1998;52:393–396. doi: 10.1111/j.1399-0039.1998.tb03062.x. [DOI] [PubMed] [Google Scholar]
- 18.Weinzierl AO, et al. Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur J Immunol. 2008;38:1503–1510. doi: 10.1002/eji.200838136. [DOI] [PubMed] [Google Scholar]
- 19.Barber LD, et al. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med. 1996;184:735–740. doi: 10.1084/jem.184.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen MH, Søndergaard I, Zeuthen J, Elliott T, Haurum JS. An assay for peptide binding to HLA-Cw*0102. Tissue Antigens. 1999;54:185–190. doi: 10.1034/j.1399-0039.1999.540210.x. [DOI] [PubMed] [Google Scholar]
- 21.Moesta AK, et al. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 22.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 23.Shimojo N, Maloy WL, Anderson RW, Biddison WE, Coligan JE. Specificity of peptide binding by the HLA-A2.1 molecule. J Immunol. 1989;143:2939–2947. [PubMed] [Google Scholar]
- 24.Kirwan SE, Burshtyn DN. Killer cell Ig-like receptor-dependent signaling by Ig-like transcript 2 (ILT2/CD85j/LILRB1/LIR-1) J Immunol. 2005;175:5006–5015. doi: 10.4049/jimmunol.175.8.5006. [DOI] [PubMed] [Google Scholar]
- 25.van Els CA, et al. A single naturally processed measles virus peptide fully dominates the HLA-A*0201-associated peptide display and is mutated at its anchor position in persistent viral strains. Eur J Immunol. 2000;30:1172–1181. doi: 10.1002/(SICI)1521-4141(200004)30:4<1172::AID-IMMU1172>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 26.Klenerman P, et al. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature. 1994;369:403–407. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 27.Bertoletti A, et al. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 28.Purbhoo MA, et al. Copresentation of natural HIV-1 agonist and antagonist ligands fails to induce the T cell receptor signaling cascade. Proc Natl Acad Sci USA. 1998;95:4527–4532. doi: 10.1073/pnas.95.8.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treanor B, et al. Microclusters of inhibitory killer immunoglobulin-like receptor signaling at natural killer cell immunological synapses. J Cell Biol. 2006;174:153–161. doi: 10.1083/jcb.200601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fricke GM, Thomas JL. Receptor aggregation by intermembrane interactions: a Monte Carlo study. Biophys Chem. 2006;119:205–211. doi: 10.1016/j.bpc.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 32.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 33.Romero V, et al. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008;45:2429–2436. doi: 10.1016/j.molimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 35.Yewdell JW, Antón LC, Bennink JR. Defective ribosomal products (DRiPs): A major source of antigenic peptides for MHC class I molecules? J Immunol. 1996;157:1823–1826. [PubMed] [Google Scholar]
- 36.Esquivel F, Yewdell J, Bennink J. RMA/S cells present endogenously synthesized cytosolic proteins to class I-restricted cytotoxic T lymphocytes. J Exp Med. 1992;175:163–168. doi: 10.1084/jem.175.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 38.Young NT, Mulder A, Cerundolo V, Claas FH, Welsh KI. Expression of HLA class I antigens in transporter associated with antigen processing (TAP)-deficient mutant cell lines. Tissue Antigens. 1998;52:368–373. doi: 10.1111/j.1399-0039.1998.tb03057.x. [DOI] [PubMed] [Google Scholar]
- 39.Winter CC, Long EO. Binding of soluble KIR-Fc fusion proteins to HLA class I. Methods Mol Biol. 2000;121:239–250. doi: 10.1385/1-59259-044-6:239. [DOI] [PubMed] [Google Scholar]
- 40.Uhrberg M, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.