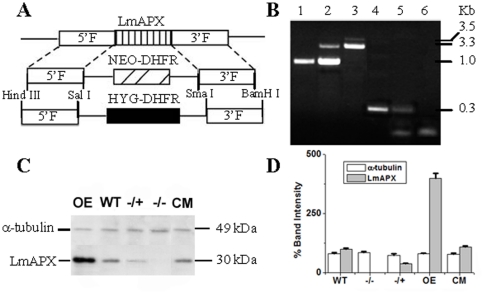
Figure 1. Targeted gene replacement of LmAPX alleles.
(A) Schematic representation of the LmAPX locus and the plasmid constructs used for gene replacement. (B) Agarose gel analysis of PCR amplified products of LmAPX gene. Lanes 1/4, 2/5 and 3/6 correspond to PCR with gDNA from WT, APX/APX::NEO heterozygous mutants and KO mutants respectively with primers external (lanes 1-3) and internal (lanes 4-6) of LmAPX gene. External forward and reverse primers were generated from 28 basepair upstream of the LmAPX gene and position 34 downstream of the stop codon of gene, respectively. The expected size of the LmAPX, NEO and HYG gene PCR product are 1.0, 3.3 and 3.5 Kb, respectively. Internal forward and reverse primers were generated from positions 360 and 623 basepair of the gene, respectively. The expected size of the PCR product from WT and APX/APX::NEO heterozygous mutants is 0.263 Kb (C) Western blot results using anti-APX and anti-α-tubulin antibody. 200 µg of L. major lysate were used for Western blotting. All the data are representative of at least three independent experiments. (D) Bar diagram depicting the % band intensities in the Western blot. Band intensity was quantified by Total Lab TL100 software (Nonlinear Dynamics Ltd) and error bars represent the SD from three independent experiments. Band intensity of wild type LmAPX was considered as a 100%. The graphs were assembled by using Origin 7.0 software (Microcal Software, Inc. Northampton, MA).