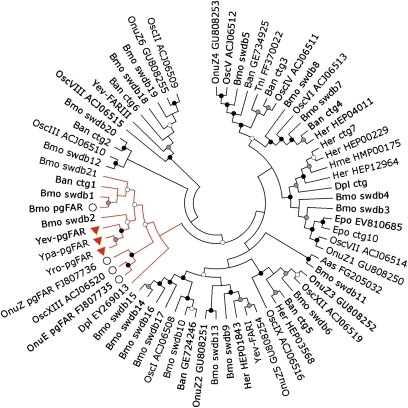
Fig. 5.
Phylogeny of Lepidopteran FARs. The Neighbor-joining algorithm analysis was computed using MEGA (v. 4.0) and the JTT model for amino acids with 1,500 bootstrap replicates (40). Nodes with bootstrap comprised between 50 and 70% are marked with open circles. Nodes with bootstrap support between 71 and 90% or over 90% are marked with gray or black circles, respectively. Lepidopteran FAR sequences were retrieved from GenBank and EST databases using BLASTP and TBLASTN searches and the B. mori pgFAR as query (16). The predicted paralogs of B. mori pgFAR were retrieved from the Silkworm Genome Database (accession numbers available in Table S1) (41). TBLASTN searches were made against the clustered ESTs database on ButterflyBase (42) and EST sequences were manually assembled into contigs (ctg) and translated using ExPASy (43). Predicted protein sequences were aligned in MAFFTv6 (44) followed by manual inspection. The clade in red contains the functional pgFARs from Y. evonymellus (Yev-pgFAR), Y. padellus (Ypa-pgFAR), and Y. rorellus (Yro-pgFAR) (upside-down red triangles). Other FAR gene members implied in pheromone biosynthesis in Lepidoptera are labeled with large open circles. The abbreviated species names correspond to (i) butterflies species: Ban, Bicyclus anynana; Dpl, Danaus plexippus; Her, Helioconius erato; Hme, Helioconius melpomene; Pxu, Papilio xuthus and (ii) moth species: Aas, Antheraea assama; Amy, Antheraea mylitta; Bmo, Bombyx mori; Epo, Epiphyas postvittana; Pin, Plodia interpunctella; Tni, Trichoplusia ni; Onu, Ostrinia nubilalis, and Osc, Ostrinia scapulalis.