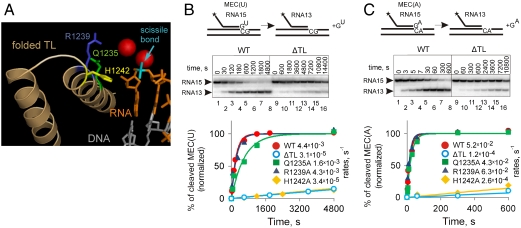
Fig. 1.
H1242 of the TL is required for hydrolysis of phosphodiester bonds. (A) Structural alignment of the yeast RNAP II elongation complex in a pretranslocated state [Protein Data Bank (PDB) ID code 1I6H (29)] and the T. thermophilus elongation complex with folded TL [PDB ID code 2O5J (7)] suggests that Q1242 (Green), R1239 (Blue), and H1242 (Yellow) of the folded TL (Light Brown) may come in close proximity to the scissile phosphodiester bond (Cyan) of RNA in the active center. RNA is shown in red and template DNA in gray. Mg2+ ions of the active center are shown as spheres. (B and C) Kinetics of phosphodiester bond hydrolysis in MEC(U) and MEC(A) (Fig. S1), respectively, for WT, ΔTL, Q1242A, R1239A, and H1242A RNAPs. Schematics above the gels show the reactions; the asterisk indicates that RNA is labeled at the 5′ end. Representative gels for WT and ΔTL are shown in the middle of each panel (gels for other mutants are shown in Fig. S2). Fractions of cleaved RNA15 against time were fitted to a single exponential equation (Solid Lines) and were normalized to the predicted amplitude. Reaction rates (s-1) are shown in the graphs legends.